The Peroxiredoxin Asp f3 Acts as Redox Sensor in Aspergillus fumigatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. A. fumigatus Strain and Culture Conditions
2.2. Construction of Fluorescent Reporter Strains
2.3. Isolation of Chromosomal DNA
2.4. Oxidative Stress Experiments
2.5. RNA Isolation
2.6. Analysis of Transcriptome Data
2.7. Gene Expression Analysis by qRT-PCR
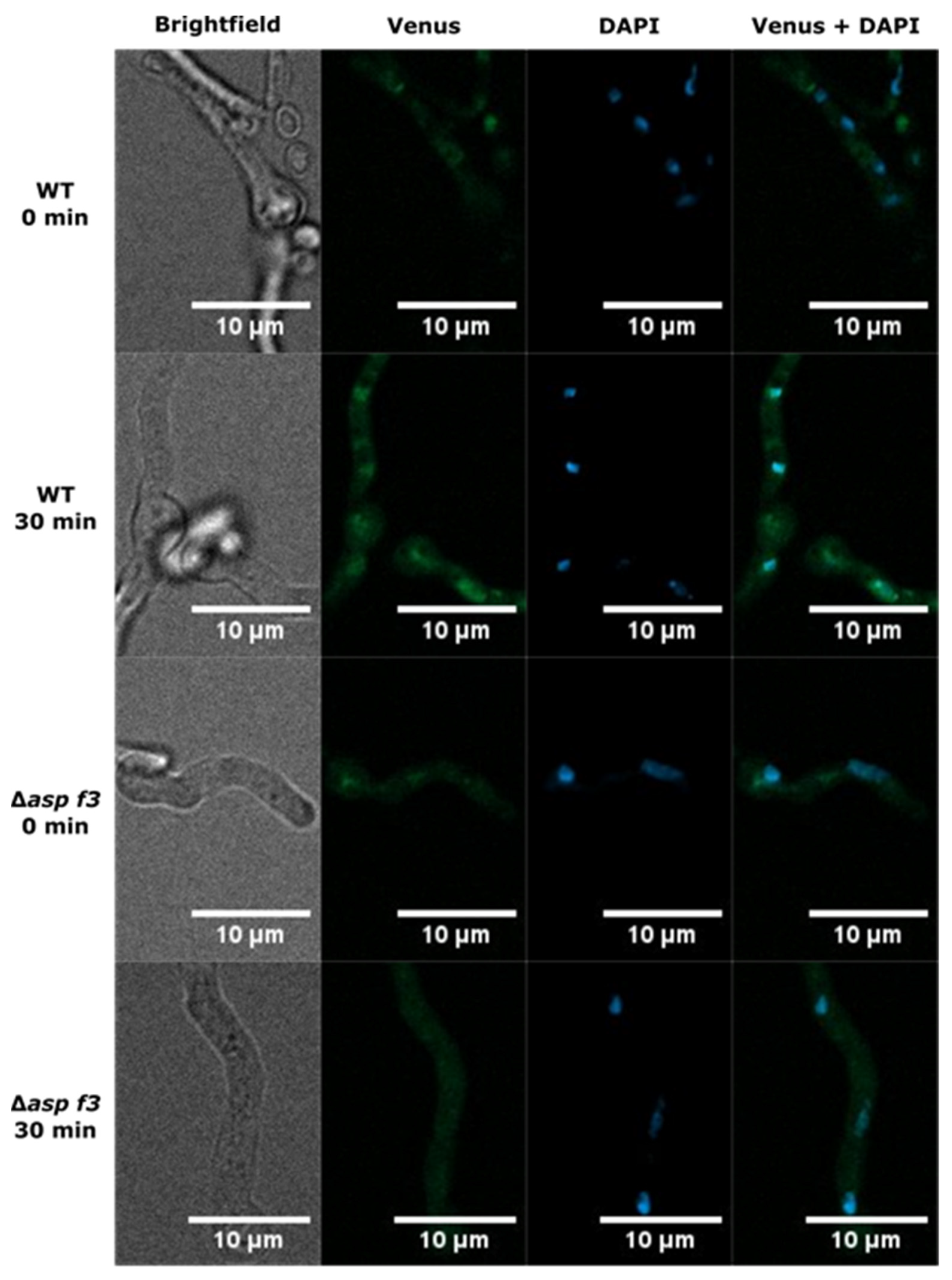
2.8. DAPI Staining and Fluorescence Microscopy
2.9. Co-Localization Analysis
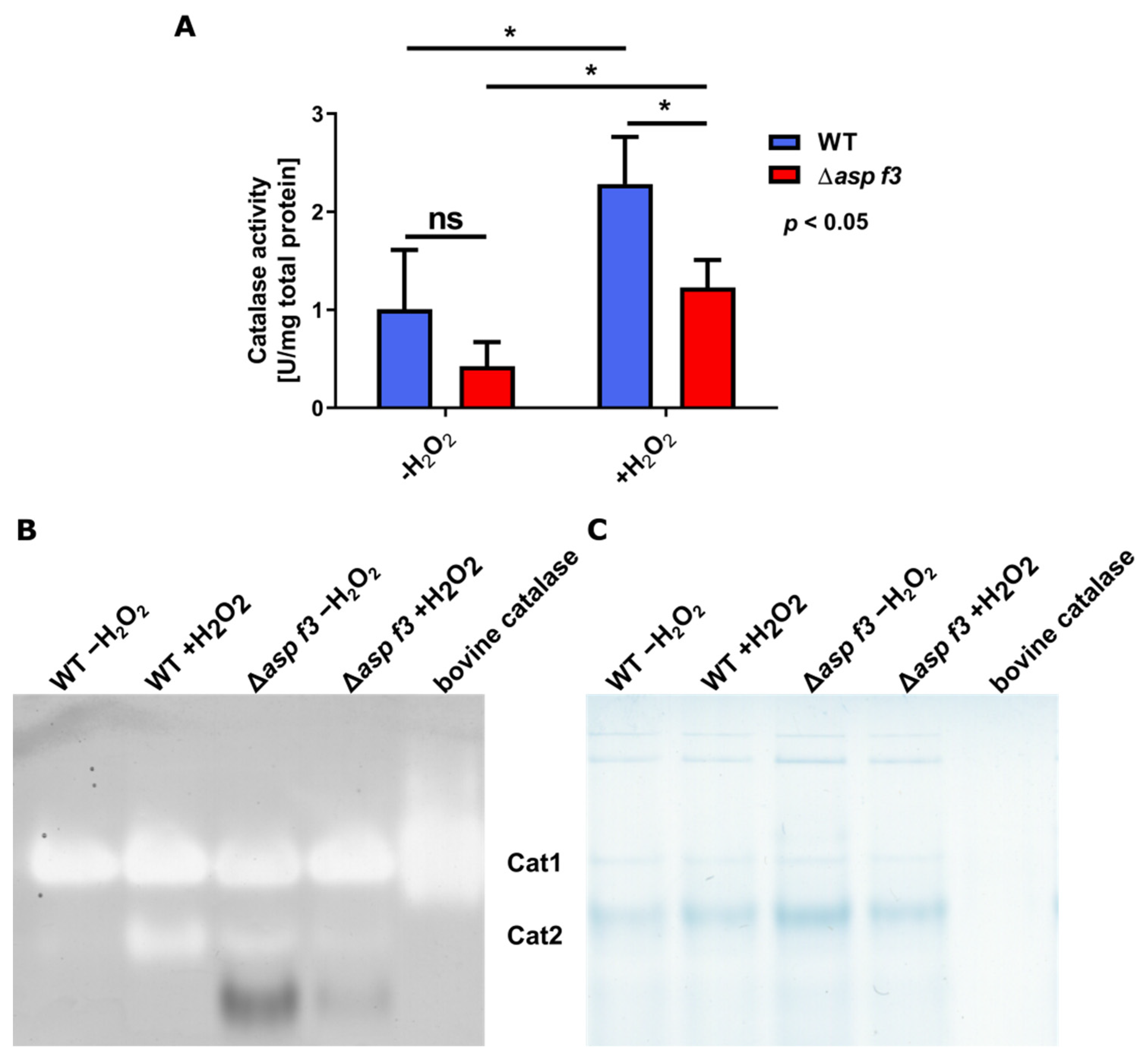
2.10. Preparation of Protein Extracts and Catalase Activity Measurements
2.11. In-Gel Catalase Activity Assay
2.12. Functional Annotation of Transcriptome Data
3. Results
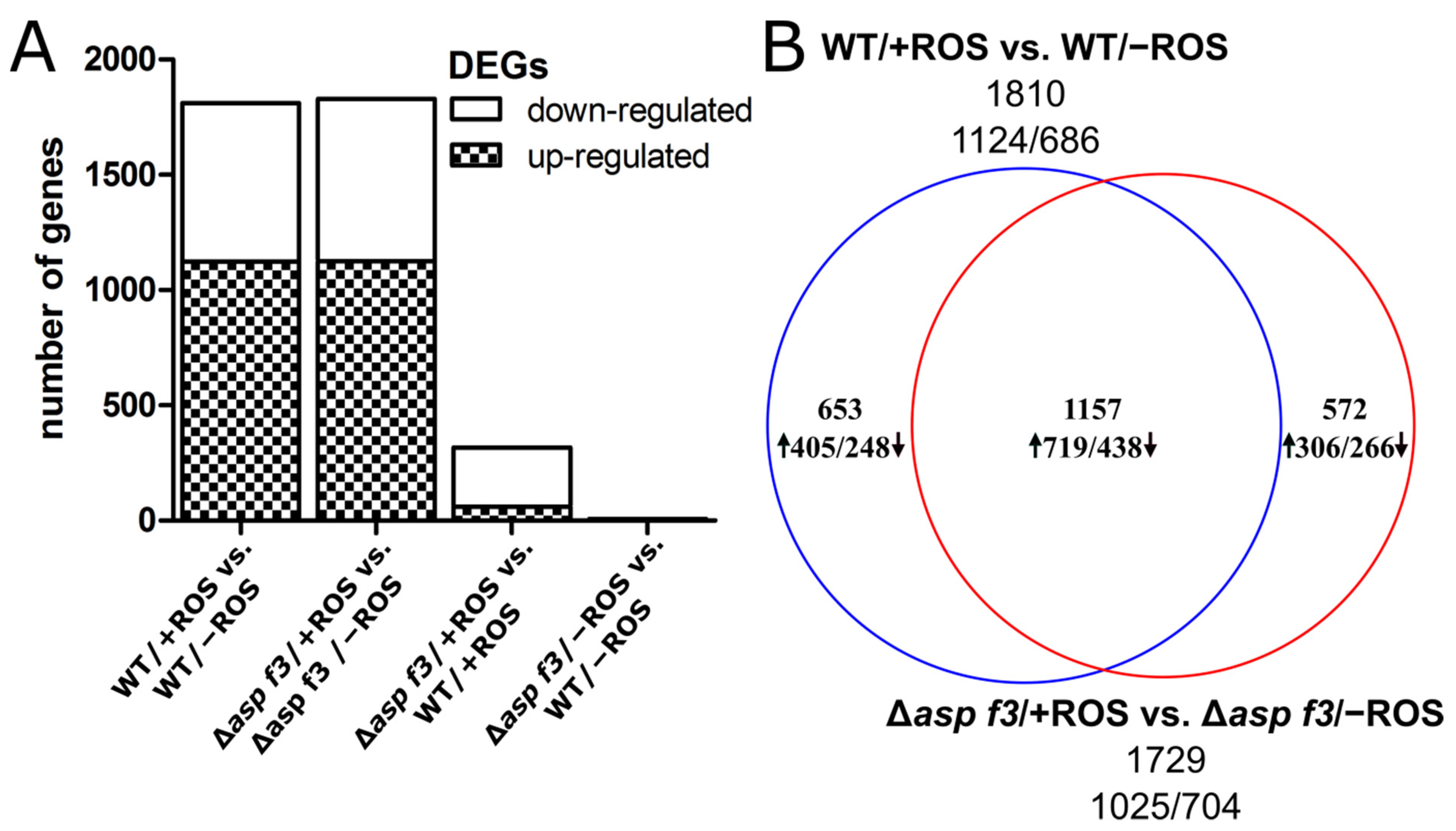
3.1. Global Transcriptome Analysis Reveals a ROS Specific Function of Asp f3
3.2. Transcriptional Induction of the ROS Defense Requires Asp f3
3.3. Full ROS Dependent Activation of Several Afyap1 Target Genes Requires Asp f3
3.4. Asp f3 Is Required for Afyap1 Activation and Nuclear Localization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brakhage, A.A.; Langfelder, K. Menacing mold: The molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 2002, 56, 433–455. [Google Scholar] [CrossRef]
- Latgé, J.-P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [CrossRef] [Green Version]
- Tekaia, F.; Latgé, J.-P. Aspergillus fumigatus: Saprophyte or pathogen? Curr. Opin. Microbiol. 2005, 8, 385–392. [Google Scholar] [CrossRef]
- Rhodes, J.C. Aspergillus fumigatus: Growth and virulence. Med. Mycol. 2006, 44, S77–S81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef] [Green Version]
- Willger, S.D.; Grahl, N.; Willger, S.D.; Grahl, N.; Cramer, R.A., Jr. Aspergillus fumigatus metabolism: Clues to mechanisms of in vivo fungal growth and virulence. Med. Mycol. 2009, 47, S72–S79. [Google Scholar] [CrossRef] [Green Version]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, rv113–rv165. [Google Scholar] [CrossRef] [Green Version]
- Asif, A.R.; Oellerich, M.; Amstrong, V.W.; Riemenschneider, B.; Monod, M.; Reichard, U. Proteome of Conidial Surface Associated Proteins of Aspergillus fumigatus Reflecting Potential Vaccine Candidates and Allergens. J. Proteome Res. 2006, 5, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.I.; Lyons, J.M.; Hong, T.B.; Tamae, D.; Liu, Y.-K.; Wilczynski, S.P.; Kalkum, M. Vaccinations with recombinant variants of Aspergillus fumigatus allergen Asp f 3 protect mice against invasive aspergillosis. Infect. Immun. 2006, 74, 5075–5084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Arevalo, D.; Bagramyan, K.; Hong, T.B.; Ito, J.I.; Kalkum, M. CD4+ T cells mediate the protective effect of the recombinant Asp f3-based anti-aspergillosis vaccine. Infect. Immun. 2011, 79, 2257–2266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillmann, F.; Bagramyan, K.; Straßburger, M.; Heinekamp, T.; Hong, T.B.; Bzymek, K.P.; Williams, J.C.; Brakhage, A.A.; Kalkum, M. The crystal structure of peroxiredoxin Asp f3 provides mechanistic insight into oxidative stress resistance and virulence of Aspergillus fumigatus. Sci. Rep. 2016, 6, 33396. [Google Scholar] [CrossRef]
- Shlezinger, N.; Irmer, H.; Dhingra, S.; Beattie, S.R.; Cramer, R.A.; Braus, G.H.; Sharon, A.; Hohl, T.M. Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death. Science 2017, 357, 1037. [Google Scholar] [CrossRef] [Green Version]
- Brakhage, A.A.; Van den Brulle, J. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J. Bacteriol. 1995, 177, 2781–2788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidner, G.; d’Enfert, C.; Koch, A.; Mol, P.C.; Brakhage, A.A. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5″-monophosphate decarboxylase. Curr. Genet. 1998, 33, 378–385. [Google Scholar] [CrossRef]
- Reichard, U.; Büttner, S.; Eiffert, H.; Staib, F.; Rüchel, R. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J. Med. Microbiol. 1990, 33, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 2009, 4, e6441. [Google Scholar] [CrossRef]
- Ballance, D.; Turner, G. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 1985, 36, 321–331. [Google Scholar] [CrossRef]
- Tilburn, J.; Sarkar, S.; Widdick, D.; Espeso, E.; Orejas, M.; Mungroo, J.; Penalva, M.; Arst, H., Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid-and alkaline-expressed genes by ambient pH. Embo J. 1995, 14, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Cenis, J. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992, 20, 2380. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 29 April 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyzes for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lessing, F.; Kniemeyer, O.; Wozniok, I.; Loeffler, J.; Kurzai, O.; Haertl, A.; Brakhage, A.A. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell 2007, 6, 2290–2302. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, I.; Hochman, A. Purification and characterization of a novel type of catalase from the bacterium Klebsiella pneumoniae. Biochim. Biophys. Acta (BBA) Gen. Subj. 1989, 991, 330–336. [Google Scholar] [CrossRef]
- Priebe, S.; Kreisel, C.; Horn, F.; Guthke, R.; Linde, J. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 2015, 31, 445–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P. The Aspergillus Genome Database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, D705–D710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paris, S.; Wysong, D.; Debeaupuis, J.-P.; Shibuya, K.; Philippe, B.; Diamond, R.D.; Latgé, J.-P. Catalases of Aspergillus fumigatus. Infect. Immun. 2003, 71, 3551–3562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, H.; Kusuya, Y.; Hagiwara, D.; Takahashi-Nakaguchi, A.; Sakai, K.; Gonoi, T. Global gene expression reveals stress-responsive genes in Aspergillus fumigatus mycelia. BMC Genom. 2017, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sugui, J.A.; Kim, H.S.; Zarember, K.A.; Chang, Y.C.; Gallin, J.I.; Nierman, W.C.; Kwon-Chung, K.J. Genes differentially expressed in conidia and hyphae of Aspergillus fumigatus upon exposure to human neutrophils. PLoS ONE 2008, 3, e2655. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Siu, K.L.; Jin, D.Y. Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J. Biol. Chem. 2004, 279, 23207–23213. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.-M.V.; Siu, K.-L.; Wong, C.-M.; Jin, D.-Y. Loss of yeast peroxiredoxin Tsa1p induces genome instability through activation of the DNA damage checkpoint and elevation of dNTP levels. PLoS Genet. 2009, 5, e1000697. [Google Scholar] [CrossRef] [Green Version]
- West, J.D.; Roston, T.J.; David, J.B.; Allan, K.M.; Loberg, M.A. Piecing together how peroxiredoxins maintain genomic stability. Antioxidants 2018, 7, 177. [Google Scholar] [CrossRef] [Green Version]
- Binder, J.; Shadkchan, Y.; Osherov, N.; Krappmann, S. The essential thioredoxin reductase of the human pathogenic mold Aspergillus fumigatus is a promising antifungal target. Front. Microbiol. 2020, 11, 1383. [Google Scholar] [CrossRef] [PubMed]
- Kupfahl, C.; Heinekamp, T.; Geginat, G.; Ruppert, T.; Hartl, A.; Hof, H.; Brakhage, A.A. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol. Microbiol. 2006, 62, 292–302. [Google Scholar] [CrossRef]
- Sugui, J.A.; Rose, S.R.; Nardone, G.; Swamydas, M.; Lee, C.-C.R.; Kwon-Chung, K.J.; Lionakis, M.S. Host immune status-specific production of gliotoxin and bis-methyl-gliotoxin during invasive aspergillosis in mice. Sci. Rep. 2017, 7, 10977. [Google Scholar] [CrossRef] [Green Version]
- Sugui, J.A.; Pardo, J.; Chang, Y.C.; Zarember, K.A.; Nardone, G.; Galvez, E.M.; Müllbacher, A.; Gallin, J.I.; Simon, M.M.; Kwon-Chung, K.J. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 2007, 6, 1562–1569. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.E.; Wiederhold, N.P.; Chi, J.; Han, X.Y.; Komanduri, K.V.; Kontoyiannis, D.P.; Prince, R.A. Detection of gliotoxin in experimental and human aspergillosis. Infect. Immun. 2005, 73, 635–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coméra, C.; André, K.; Laffitte, J.; Collet, X.; Galtier, P.; Maridonneau-Parini, I. Gliotoxin from Aspergillus fumigatus affects phagocytosis and the organization of the actin cytoskeleton by distinct signalling pathways in human neutrophils. Microbes Infect. 2007, 9, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer, R.A.; Gamcsik, M.P.; Brooking, R.M.; Najvar, L.K.; Kirkpatrick, W.R.; Patterson, T.F.; Balibar, C.J.; Graybill, J.R.; Perfect, J.R.; Abraham, S.N. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 2006, 5, 972–980. [Google Scholar] [CrossRef] [Green Version]
- Hof, H.; Kupfahl, C. Gliotoxin in Aspergillus fumigatus: An example that mycotoxins are potential virulence factors. Mycotoxin Res. 2009, 25, 123. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, L.; Owens, R.A.; Dolan, S.K.; O’Keeffe, G.; Schrettl, M.; Kavanagh, K.; Jones, G.W.; Doyle, S. The Aspergillus fumigatus protein GliK protects against oxidative stress and is essential for gliotoxin biosynthesis. Eukaryot. Cell 2012, 11, 1226–1238. [Google Scholar] [CrossRef] [Green Version]
- Delaunay, A.; Pflieger, D.; Barrault, M.-B.; Vinh, J.; Toledano, M.B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 2002, 111, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Martínez, A.E.; Cano-Domínguez, N.; Aguirre, J. Yap1 homologs mediate more than the redox regulation of the antioxidant response in filamentous fungi. Fungal Biol. 2020, 124, 253–262. [Google Scholar] [CrossRef]
- Mendoza-Martínez, A.E.; Lara-Rojas, F.; Sánchez, O.; Aguirre, J. NapA Mediates a Redox Regulation of the Antioxidant Response, Carbon Utilization and Development in Aspergillus nidulans. Front. Microbiol. 2017, 8, 516. [Google Scholar] [CrossRef]
- Okazaki, S.; Naganuma, A.; Kuge, S. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid Redox Signal. 2005, 7, 327–334. [Google Scholar] [CrossRef]
- Park, S.G.; Cha, M.K.; Jeong, W.; Kim, I.H. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 5723–5732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Yu, H.; Zhou, Z.; Takaya, N.; Zhou, S.; Wang, P. Peroxiredoxin System of Aspergillus nidulans Resists Inactivation by High Concentration of Hydrogen Peroxide-Mediated Oxidative Stress. J. Microbiol. Biotechnol. 2018, 28, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Kurucz, V.; Krüger, T.; Antal, K.; Dietl, A.-M.; Haas, H.; Pócsi, I.; Kniemeyer, O.; Emri, T. Additional oxidative stress reroutes the global response of Aspergillus fumigatus to iron depletion. BMC Genom. 2018, 19, 357. [Google Scholar] [CrossRef] [PubMed]




| Strain | Genotype | References |
|---|---|---|
| A. fumigatus D141 | WT | [15] |
| A. fumigatus Δasp f3 | Asp f3::hph; HygR | [11] |
| A. fumigatus Δasp f3C | Asp f3::hph; HygR Δasp f3::Asp f3; PTR | [11] |
| A. fumigatus OE::Afyap1VENUS | PGpdA-Afyap1Venus-Tnos::ptrA; PTR | This study |
| A. fumigatus Δasp f3 OE::Afyap1VENUS | Asp f3::hph; HygR PGpdA-Afyap1Venus-Tnos::ptrA; PTR | This study |
| Gene ID | Afyap1 Target * | WT + ROS vs. WT − ROS | Δasp f3 + ROS vs. Δasp f3 − ROS | Δasp f3 + ROS vs. WT + ROS | |
|---|---|---|---|---|---|
| 1 | Putative NADH flavin oxidoreductase (AFUA_2g04060) | − | 4.16 | 1.93 | −2.65 |
| 2 | bifunctional catalase-peroxidase (cat2, AFUA_8g01670) | + | 2.85 | 0.31 | −2.64 |
| 3 | p-Nitroreductase family protein (pnr1, AFUA_5g09910) | + | 4.05 | 2.4 | −2.15 |
| 4 | Oxidoreductase, putative (AFUA_5G01250) | − | 2.12 | 0.39 | −1.87 |
| 5 | Thioredoxin reductase (trxR, AFUA_4g12990) | − | 2.75 | 1.79 | −1.74 |
| 6 | Glutathione transferase, putative (AFUA_2g15770) | − | 2.53 | 0.95 | −1.66 |
| 7 | NADH-dependent flavin oxidoreductase, putative (AFUA_7G06420) | − | 1.81 | 0.88 | −1.47 |
| 8 | Glutathione peroxidase (gpx3, AFUA_3g12270) | − | 1.31 | 0.35 | −1.34 |
| 9 | Cytochrome c peroxidase (ccp1, AFUA_4G09110) | + | 1.36 | 0.21 | −1.3 |
| 10 | Glutathione S-transferase, putative (AFUA_2G00590) | − | 0.85 | 0.22 | −1.18 |
| 11 | Ferric-chelate reductase, putative (AFUA_6G13750) | − | 0.86 | −0.26 | −1.14 |
| 12 | Gliotoxin Cluster e.G. gliM (AFUA_6G09680) | − | −0.3 | 0.62 | −1.12 |
| 13 | Metalloreductase, putative (AFUA_6g02820) | − | 1.21 | 0.27 | −1.1 |
| 14 | Thioredoxin (Asp29/Trx1) (AFUA_5g11320) | − | 1.63 | 0.7 | −1.1 |
| 15 | Mitochondrial peroxiredoxin Prx1 (AFUA_4G08580) | + | −0.75 | −0.66 | 0.13 |
| 16 | Methionine synthase MetH/D (AFUA_4G07360) | + | −0.11 | −0.02 | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boysen, J.M.; Saeed, N.; Wolf, T.; Panagiotou, G.; Hillmann, F. The Peroxiredoxin Asp f3 Acts as Redox Sensor in Aspergillus fumigatus. Genes 2021, 12, 668. https://doi.org/10.3390/genes12050668
Boysen JM, Saeed N, Wolf T, Panagiotou G, Hillmann F. The Peroxiredoxin Asp f3 Acts as Redox Sensor in Aspergillus fumigatus. Genes. 2021; 12(5):668. https://doi.org/10.3390/genes12050668
Chicago/Turabian StyleBoysen, Jana Marie, Nauman Saeed, Thomas Wolf, Gianni Panagiotou, and Falk Hillmann. 2021. "The Peroxiredoxin Asp f3 Acts as Redox Sensor in Aspergillus fumigatus" Genes 12, no. 5: 668. https://doi.org/10.3390/genes12050668






