Changes in Allele Frequencies When Different Genomic Coancestry Matrices Are Used for Maintaining Genetic Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of the Base Population
2.2. Management Strategies
2.3. Parameters Evaluated
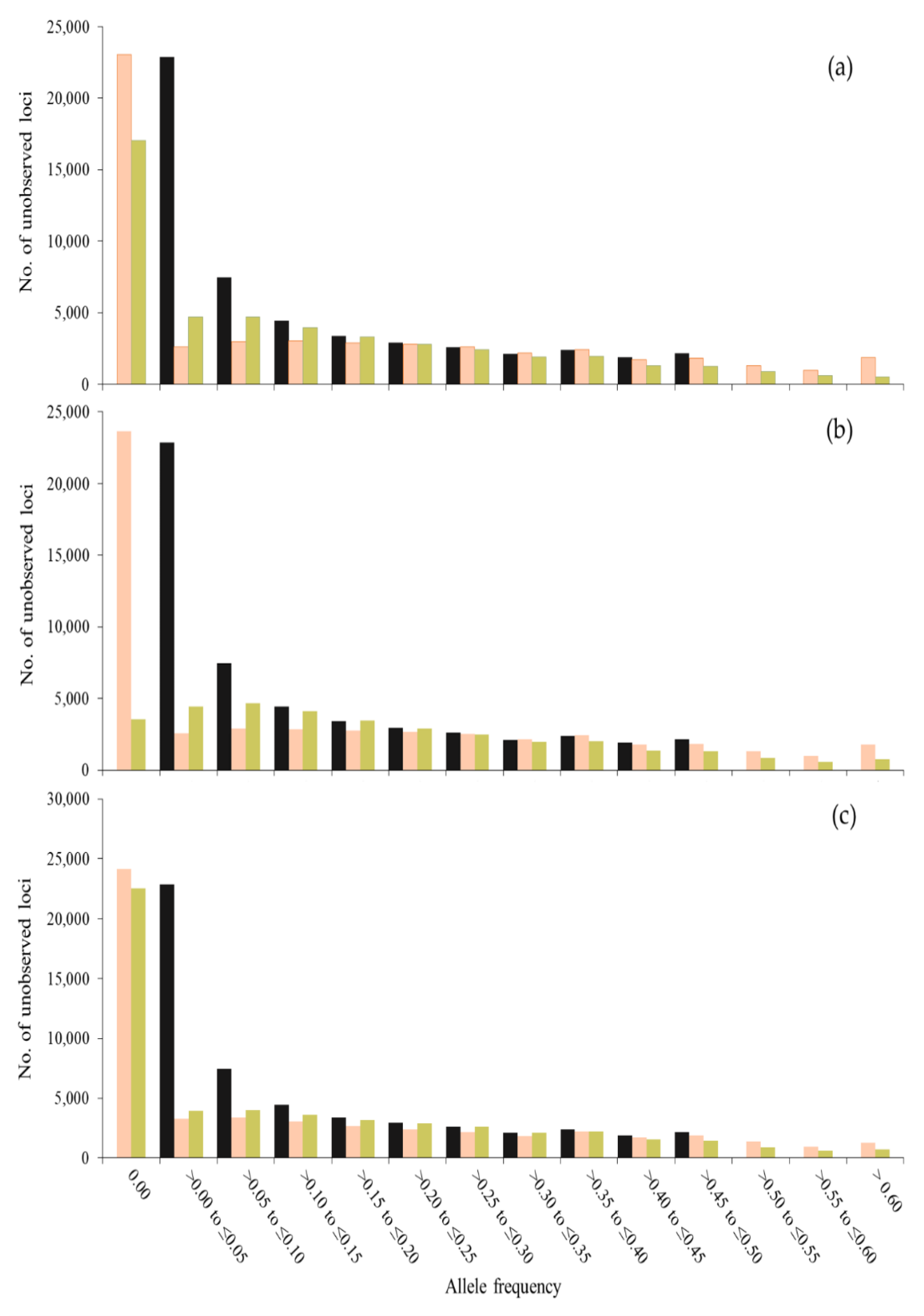
3. Results
3.1. Expected Heterozygosity and Kullback–Leibler Divergence for Populations of Size N = 100
3.2. Expected Heterozygosity and Kullback–Leibler Divergence for Populations of Size N = 20
3.3. Effective Population Size
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; Ballou, J.D. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Meuwissen, T. Maximizing the response of selection with a predefined rate of inbreeding. J. Anim. Sci. 1997, 75, 934–940. [Google Scholar] [CrossRef]
- Grundy, B.; Villanueva, B.; Woolliams, J.A. Dynamic selection procedures for constrained inbreeding and their consequences for pedigree development. Genet. Res. 1998, 72, 159–168. [Google Scholar] [CrossRef]
- Fernández, J.; A Toro, M.; Caballero, A. Fixed contributions designs vs. minimization of global coancestry to control inbreeding in small populations. Genetics 2003, 165, 885–894. [Google Scholar] [CrossRef]
- Caballero, A.; Toro, M.A. Interrelations between effective population size and other pedigree tools for the management of conserved populations. Genet. Res. 2000, 75, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S.; Mackay, T.F.C. Introduction to quantitative genetics. In Introduction to Quantitative Genetics, 4th ed.; Longman: Harlow, UK, 1996. [Google Scholar]
- Lacy, R.C. Should we select genetic alleles in our conservation breeding programs? Zoo Biol. 2000, 19, 279–282. [Google Scholar] [CrossRef]
- Frankham, R. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 2008, 17, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Saura, M.; Pérez-Figueroa, A.; Fernández, J.; Toro, M.A.; Caballero, A. Preserving population allele frequencies in ex situ conservation programs. Conserv. Biol. 2008, 22, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- De Cara, M.A.R.; Fernández, J.; Toro, M.A.; Villanueva, B. Using genome-wide information to minimize the loss of diversity in conservation programs. J. Anim. Breed. Genet. 2011, 128, 456–464. [Google Scholar] [CrossRef]
- De Cara, M.; Ángeles, R.; Villanueva, B.; Toro, M.Á.; Fernández, J. Using genomic tools to maintain diversity and fitness in conservation programmes. Mol. Ecol. 2013, 22, 6091–6099. [Google Scholar] [CrossRef]
- Gómez-Romano, F.; Villanueva, B.; De Cara, M.Á.R.; Fernández, J. Maintaining genetic diversity using molecular coancestry: The effect of marker density and effective population size. Genet. Sel. Evol. 2013, 45, 38. [Google Scholar] [CrossRef] [Green Version]
- Fernández, J.; Toro, M.A.; Caballero, A. Managing Individuals’ Contributions to Maximize the Allelic Diversity Maintained in Small, Conserved Populations. Conserv. Biol. 2004, 18, 1358–1367. [Google Scholar] [CrossRef]
- De Cara, M.A.R.; Villanueva, B.; Toro, M.A.; Fernández, J. Purging deleterious mutations in conservation programs: Combining optimal contributions with inbred mattings. Heredity 2013, 110, 530–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eynard, S.E.; Windig, J.J.; Hiemstra, S.J.; Calus, M.P.L. Whole-genome sequence data uncover loss of genetic diversity due to selection. Genet. Sel. Evol. 2016, 48, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales-González, E.; Saura, M.; Fernández, A.; Fernández, J.; Pong-Wong, R.; Cabaleiro, S.; Martínez, P.; Martín-García, A.; Villanueva, B. Evaluating different genomic coancestry matrices for managing genetic variability in turbot. Aquaculture 2020, 520, 734985. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Sonesson, A.K.; Gebregiwergis, G.; Woolliams, J.A. Management of Genetic Diversity in the Era of Genomics. Front. Genet. 2020, 11, 880. [Google Scholar] [CrossRef]
- Li, C.C.; Horvitz, D.G. Some methods of estimating the inbreeding coefficient. Am. J. Hum. Genet. 1953, 5, 107–117. [Google Scholar]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.D.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Romano, F.; Villanueva, B.; Fernández, J.; Woolliams, J.A.; Pong-Wong, R. The use of genomic coancestry matrices in the optimisation of contributions to maintain genetic diversity at specific regions of the genome. Genet. Sel. Evol. 2016, 48, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Nejati-Javaremi, A.; Smith, C.; Gibson, J.P. Effect of total allelic relationship on accuracy of evaluation and response to selection. J. Anim. Sci. 1997, 75, 1738–1745. [Google Scholar] [CrossRef]
- Toro, M.A.; Villanueva, B.; Fernández, J. The concept of effective population size loses its meaning in the context of optimal management of diversity using molecular markers. J. Anim. Breed. Genet. 2019, 137, 345–355. [Google Scholar] [CrossRef]
- Woolliams, J.A.; Berg, P.; Dagnachew, B.S.; Meuwissen, T.H.E. Genetic contributions and their optimisation. J. Anim. Breed. Genet. 2015, 132, 89–99. [Google Scholar] [CrossRef]
- Toro, M.; Barragán, C.; Óvilo, C.; Rodrigañez, J.; Rodriguez, C.; Silió, L. Estimation of coancestry in Iberian pigs using molecular markers. Conserv. Genet. 2002, 3, 309–320. [Google Scholar] [CrossRef]
- Forni, S.; Aguilar, I.; Misztal, I. Different genomic relationship matrices for single-step analysis using phenotypic, pedigree and genomic information. Genet. Sel. Evol. 2011, 43, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullback, S. Information Theory and Statistics; Dover Publications: Mineola, New York, NY, USA, 1997. [Google Scholar]
- Fernández, J.; Caballero, A. Accumulation of deleterious mutations and equalization of parental contributions in the conservation of genetic resources. Heredity 2001, 86, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, B.; Fernández, A.; Saura, M.; Caballero, A.; Fernández, J. The value of genomic relationship matrices for estimating inbreeding. Genet. Sel. Evol. 2021. under review. [Google Scholar]
- Fernã¡ndez, J.; Toro, M.Ã.; Sonesson, A.K.; Villanueva, B.; Fernández, J. Optimizing the creation of base populations for aquaculture breeding programs using phenotypic and genomic data and its consequences on genetic progress. Front. Genet. 2014, 5, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, J.; Roughsedge, T.; Woolliams, J.A.; Villanueva, B. Optimization of the sampling strategy for establishing a gene bank: Storing PrP alleles following a scrapie eradication plan as a case study. Anim. Sci. 2006, 82, 813–821. [Google Scholar] [CrossRef]
- Sonesson, A.K.; Janss, L.L.; Meuwissen, T.H. Selection against genetic defects in conservation schemes while controlling inbreeding. Genet. Sel. Evol. 2003, 35, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Coppieters, W.; Rollin, F.; Desmecht, D.; Agerholm, J.S.; Cambisano, N.; Carta, E.; Dardano, S.; Dive, M.; Fasquelle, C.; et al. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat. Genet. 2008, 40, 449–454. [Google Scholar] [CrossRef]
- Caballero, A.; Rodríguez-Ramilo, S.T. A new method for the partition of allelic diversity within and between subpopulations. Conserv. Genet. 2010, 11, 2219–2229. [Google Scholar] [CrossRef]
- James, J.W. The founder effect and response to artificial selection. Genet. Res. 1970, 16, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Rasbash, J. Models of long term artificial selection in finite population. Genet. Res. 1986, 48, 41–50. [Google Scholar] [CrossRef] [PubMed]


| SE | SO_LH * | SO_VR * | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t | He | KL | NS | S | U | He | KL | NS | S | U | He | KL | NS | S | U | ||
| 1 | 19.17 | 0.06 | 100 | 51,035 | 50,894 | +0.14 | +0.14 | −39 | −2239 | −2246 | 0.00 | 0.00 | 0 | +8 | +18 | ||
| 2 | 19.12 | 0.12 | 100 | 49,873 | 49,737 | +0.21 | +0.23 | −36 | −3206 | −3229 | 0.00 | 0.00 | 0 | −22 | 0 | ||
| 3 | 19.07 | 0.18 | 100 | 48,852 | 48,729 | +0.28 | +0.30 | −35 | −3792 | −3847 | 0.00 | 0.00 | 0 | −61 | −52 | ||
| 4 | 19.03 | 0.24 | 100 | 47,946 | 47,828 | +0.35 | +0.37 | −35 | −4182 | −4261 | 0.00 | 0.00 | −1 | −113 | −101 | ||
| 5 | 18.98 | 0.30 | 100 | 47,108 | 47,003 | +0.41 | +0.43 | −33 | −4384 | −4499 | 0.00 | −0.01 | −1 | −162 | −157 | ||
| 10 | 18.73 | 0.57 | 100 | 43,777 | 43,691 | +0.68 | +0.68 | −30 | −4731 | −4975 | 0.00 | −0.03 | −2 | −399 | −401 | ||
| 15 | 18.51 | 0.82 | 100 | 41,311 | 41,217 | +0.89 | +0.86 | −28 | −4523 | −4855 | −0.01 | −0.06 | −5 | −595 | −587 | ||
| 20 | 18.27 | 1.06 | 100 | 39,313 | 39,229 | +1.08 | +0.99 | −26 | −4152 | −4567 | −0.01 | −0.09 | −6 | −714 | −720 | ||
| 30 | 17.82 | 1.50 | 100 | 36,231 | 36,140 | +1.40 | +1.16 | −24 | −3329 | −3896 | +0.01 | −0.18 | −9 | −906 | −899 | ||
| 40 | 17.38 | 1.90 | 100 | 33,854 | 33,759 | +1.67 | +1.24 | −22 | −2517 | −3215 | +0.03 | −0.26 | −11 | −995 | −970 | ||
| 50 | 16.95 | 2.28 | 100 | 31,940 | 31,848 | +1.92 | +1.27 | −21 | −1786 | −2594 | +0.05 | −0.35 | −12 | −1081 | −1036 | ||
| SNPs | Unobserved Loci | |||||||
|---|---|---|---|---|---|---|---|---|
| t | SE | SO_LH | SO_VR | SE | SO_LH | SO_VR | ||
| 0 | 13.45 | 13.45 | 13.45 | 13.39 | 13.39 | 13.39 | ||
| 1 | 13.44 | 13.68 | 13.45 | 13.39 | 13.60 | 13.40 | ||
| 2 | 13.44 | 13.81 | 13.45 | 13.39 | 13.72 | 13.40 | ||
| 3 | 13.44 | 13.94 | 13.45 | 13.38 | 13.82 | 13.39 | ||
| 4 | 13.44 | 14.06 | 13.44 | 13.38 | 13.93 | 13.39 | ||
| 5 | 13.44 | 14.17 | 13.44 | 13.38 | 14.02 | 13.39 | ||
| 10 | 13.44 | 14.67 | 13.41 | 13.38 | 14.44 | 13.36 | ||
| 15 | 13.45 | 15.08 | 13.37 | 13.39 | 14.77 | 13.33 | ||
| 20 | 13.44 | 15.42 | 13.32 | 13.39 | 15.05 | 13.29 | ||
| 30 | 13.44 | 15.96 | 13.23 | 13.39 | 15.46 | 13.23 | ||
| 40 | 13.45 | 16.36 | 13.12 | 13.39 | 15.75 | 13.15 | ||
| 50 | 13.45 | 16.67 | 13.01 | 13.40 | 15.98 | 13.07 | ||
| SE | SO_LH * | SO_VR * | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t | He | KL | NS | S | U | He | KL | NS | S | U | He | KL | NS | S | U | ||
| 1 | 23.35 | 0.27 | 20 | 38,995 | 38,955 | +0.04 | +0.05 | −1 | −193 | −233 | +0.03 | 0.00 | 0 | +31 | +134 | ||
| 2 | 23.06 | 0.52 | 20 | 37,093 | 37,050 | +0.06 | +0.07 | −1 | −275 | −335 | +0.01 | 0.00 | 0 | +52 | +155 | ||
| 3 | 22.76 | 0.76 | 20 | 35,522 | 35,472 | +0.10 | +0.09 | −1 | −356 | −410 | −0.02 | +0.01 | 0 | −12 | +104 | ||
| 4 | 22.48 | 0.99 | 20 | 34,166 | 34,119 | +0.07 | +0.11 | −1 | −390 | −442 | −0.02 | −0.01 | 0 | −16 | +94 | ||
| 5 | 22.19 | 1.20 | 20 | 33,016 | 32,978 | +0.08 | +0.13 | −1 | −456 | −528 | −0.03 | 0.00 | 0 | −69 | +37 | ||
| 10 | 20.79 | 2.17 | 20 | 28,782 | 28,692 | +0.17 | +0.18 | −1 | −533 | −563 | −0.07 | −0.03 | −1 | −269 | −62 | ||
| 15 | 19.52 | 3.00 | 20 | 25,844 | 25,763 | +0.24 | +0.17 | −1 | −497 | −563 | −0.03 | −0.07 | −1 | −400 | −206 | ||
| 20 | 18.33 | 3.75 | 20 | 23,512 | 23,434 | +0.37 | +0.13 | −1 | −336 | −424 | −0.01 | −0.12 | −1 | −429 | −247 | ||
| 30 | 16.02 | 5.13 | 20 | 19,854 | 19,795 | +0.79 | −0.02 | −2 | +81 | −59 | +0.04 | −0.25 | −2 | −469 | −337 | ||
| 40 | 14.03 | 6.26 | 20 | 17,044 | 17,002 | +1.15 | −0.16 | −1 | +545 | +377 | +0.18 | −0.43 | −2 | −432 | −309 | ||
| 50 | 12.32 | 7.23 | 20 | 14,853 | 14,811 | +1.39 | −0.27 | −1 | +787 | +592 | +0.19 | −0.52 | −2 | −433 | −322 | ||
| N = 100 | N = 20 | ||||||
|---|---|---|---|---|---|---|---|
| t | SE | SO_LH | SO_VR | SE | SO_LH | SO_VR | |
| 1 | 188.21 | −111.90 | 195.55 | 36.92 | 42.27 | 40.40 | |
| 5 | 199.07 | −855.78 | 197.46 | 36.78 | 41.24 | 34.31 | |
| 10 | 191.56 | −5777.32 | 193.05 | 38.54 | 40.81 | 41.77 | |
| 15 | 203.50 | 1855.71 | 194.54 | 36.65 | 45.41 | 43.18 | |
| 20 | 202.62 | 1033.03 | 201.52 | 40.61 | 47.25 | 40.02 | |
| 25 | 190.44 | 636.00 | 209.85 | 40.20 | 47.08 | 42.02 | |
| 30 | 193.58 | 670.07 | 209.79 | 36.45 | 53.03 | 38.57 | |
| 35 | 193.30 | 524.97 | 206.03 | 33.41 | 50.28 | 44.62 | |
| 40 | 204.95 | 601.67 | 212.53 | 36.94 | 47.91 | 49.68 | |
| 45 | 207.44 | 703.31 | 205.00 | 37.52 | 48.50 | 40.09 | |
| 50 | 206.86 | 481.08 | 213.02 | 41.99 | 46.20 | 38.53 | |
| SO_LH | SO_VR | |||||||
|---|---|---|---|---|---|---|---|---|
| t | S | NS | ESf | Sf | NS | ESf | Sf | |
| 1 | 20 | 7 | 0.3 | 0 | 20 | 0.3 | 0 | |
| 2 | 7 | 0.7 | 0 | 13 | 0.8 | 1 | ||
| 3 | 8 | 0.8 | 0 | 13 | 1.4 | 1 | ||
| 4 | 8 | 0.9 | 0 | 12 | 1.7 | 1 | ||
| 1 | 1000 | 15 | 21.7 | 21 | 20 | 17.6 | 18 | |
| 2 | 16 | 38.9 | 37 | 19 | 34.6 | 33 | ||
| 3 | 15 | 54.6 | 52 | 19 | 50.9 | 47 | ||
| 4 | 15 | 68.6 | 64 | 18 | 66.3 | 60 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-González, E.; Fernández, J.; Pong-Wong, R.; Toro, M.Á.; Villanueva, B. Changes in Allele Frequencies When Different Genomic Coancestry Matrices Are Used for Maintaining Genetic Diversity. Genes 2021, 12, 673. https://doi.org/10.3390/genes12050673
Morales-González E, Fernández J, Pong-Wong R, Toro MÁ, Villanueva B. Changes in Allele Frequencies When Different Genomic Coancestry Matrices Are Used for Maintaining Genetic Diversity. Genes. 2021; 12(5):673. https://doi.org/10.3390/genes12050673
Chicago/Turabian StyleMorales-González, Elisabeth, Jesús Fernández, Ricardo Pong-Wong, Miguel Ángel Toro, and Beatriz Villanueva. 2021. "Changes in Allele Frequencies When Different Genomic Coancestry Matrices Are Used for Maintaining Genetic Diversity" Genes 12, no. 5: 673. https://doi.org/10.3390/genes12050673






