The Adenine/Thymine Deleterious Selection Model for GC Content Evolution at the Third Codon Position of the Histone Genes in Drosophila
Abstract
:1. Introduction
2. Evolution of the GC Content at the Third Codon Position of the Histone Genes in Drosophila
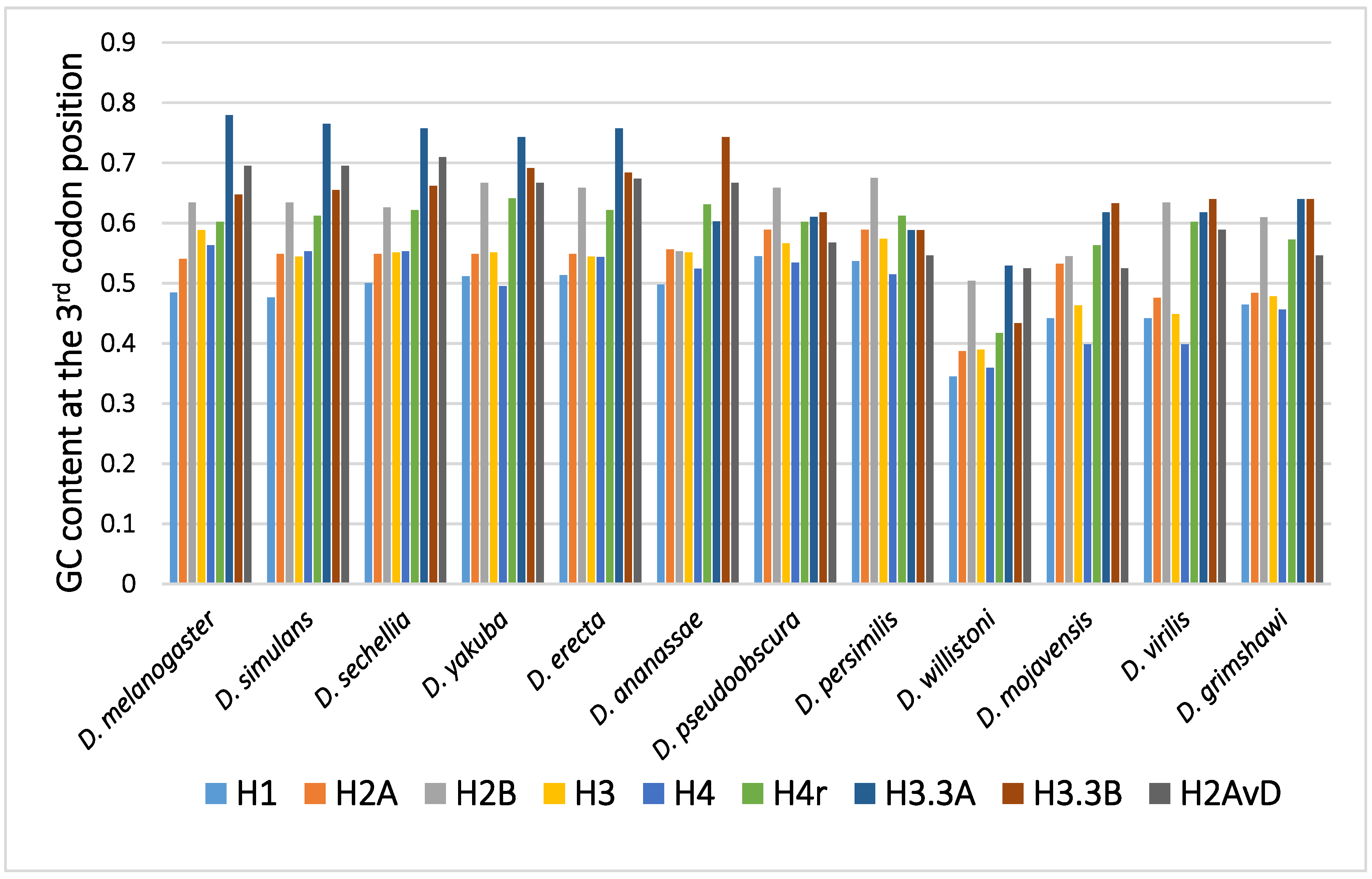
2.1. Disparity in the GC Content at the Third Codon Position among the Genes
2.2. Disparity in the GC Content at the Third Codon Position between the Genes of the Canonical and Replacement Types of Histones
2.3. Disparity in GC Content at the Third Codon Position of the Genes among the Different Species
2.4. Mode of the Evolution of GC Content at the Third Codon Position According to Phylogeny
3. Models for the Evolution of GC Content
3.1. Mutation Bias
3.2. Deleterious Selection for A/T
3.3. Effect of the Population Size
4. Generality of the Model
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kornberg, R.D. Chromatin Structure: A Repeating Unit of Histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Kornberg, R.D. Structure of Chromatin. Annu. Rev. Biochem. 1977, 46, 931–954. [Google Scholar] [CrossRef]
- McGhee, J.D.; Rau, D.C.; Charney, E.; Felsenfeld, G. Orientation of the nucleosome within the higher order structure of chromatin. Cell 1980, 22, 87–96. [Google Scholar] [CrossRef]
- Cutter, A.R.; Hayes, J.J. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef] [Green Version]
- Schümperli, D. Cell-cycle regulation of histone gene expression. Cell 1986, 45, 471–472. [Google Scholar] [CrossRef]
- Malik, H.S.; Henikoff, S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 2001, 157, 1293–1298. [Google Scholar] [CrossRef]
- Vermaak, D.; Hayden, H.S.; Henikoff, S. Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 2002, 22, 7553–7561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalal, Y.; Furuyama, T.; Vermaak, D.; Henikoff, S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 15974–15981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, D.; Snyder, L.A.; Blumenfeld, M. Drosophila nucleosomes contain an unusual histone-like protein. Proc. Natl. Acad. Sci. USA 1980, 77, 2671–2675. [Google Scholar] [CrossRef] [Green Version]
- Lifton, R.P.; Goldberg, M.L.; Karp, R.W.; Hogness, D.S. The Organization of the Histone Genes in Drosophila melanogaster: Functional and Evolutionary Implications. Cold Spring Harb. Symp. Quant. Biol. 1978, 42, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Pardue, M.L.; Kedes, L.H.; Weinberg, E.S.; Birnstiel, M.L. Localization of sequences coding for histone messenger RNA in the chromosomes of Drosophila melanogaster. Chromosoma 1977, 63, 135–151. [Google Scholar] [CrossRef]
- Fretzin, S.; Allan, B.D.; van Daal, A.; Elgin, S.C.R. A Drosophila melanogaster H3.3 cDNA encodes a histone variant identical with the vertebrate H3.3. Gene 1991, 107, 341–342. [Google Scholar] [CrossRef]
- Van Daal, A.; White, E.M.; Gorovsky, M.A.; Elgin, C.R. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A. F/Z type. Nucleic Acids Res. 1988, 16, 7487–7497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhmanova, A.S.; Bindels, P.C.T.; Xu, J.; Miedema, K.; Kremer, H.; Hennig, W.; Xu, J.; Hennig, W. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome 1995, 38, 586–600. [Google Scholar] [CrossRef]
- Akhmanova, A.; Miedema, K.; Hennig, W. Identification and characterization of the Drosophila histone H4 replacement gene. FEBS Lett. 1996, 388, 219–222. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [Green Version]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [Green Version]
- Musselman, C.A.; Lalonde, M.-E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Benson, L.J.; Gu, Y.; Yakovleva, T.; Tong, K.; Barrows, C.; Strack, C.L.; Cook, R.G.; Mizzen, C.A.; Annunziato, A.T. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 2006, 281, 9287–9296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef]
- Santoro, S.W.; Dulac, C. Histone variants and cellular plasticity. Trends Genet. 2015, 31, 516–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biterge, B.; Schneider, R. Histone variants: Key players of chromatin. Cell Tissue Res. 2014, 356, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Maze, I.; Noh, K.-M.; Soshnev, A.A.; Allis, C.D. Every amino acid matters: Essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 2014, 15, 259–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, P.B.; Workman, J.L. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 2013, 5, a017905. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef]
- Gaume, X.; Torres-Padilla, M.-E. Regulation of reprogramming and cellular plasticity through histone exchange and histone variant incorporation. Cold Spring Harb. Symp. Quant. Biol. 2015, 80, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Henikoff, S. Epigenetic profiling of histone variants. Epigenomics 2009, 101–118. [Google Scholar] [CrossRef]
- Matsuo, Y.; Yamazaki, T. tRNA derived insertion element in histone gene repeating unit of Drosophila melanogaster. Nucleic Acids Res. 1989, 17, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, Y. Genomic structure and evolution of the histone gene family in Drosophila. Curr. Top. Genet. 2006, 2, 1–14. [Google Scholar]
- Matsuo, Y.; Yamazaki, T. Nucleotide variation and divergence in the histone multigene family in Drosophila melanogaster. Genetics 1989, 122, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.; Bains, W.; Kedes, L. Codon usage in histone gene families of higher eukaryotes reflects functional rather than phylogenetic relationships. J. Mol. Evol. 1986, 23, 224–241. [Google Scholar] [CrossRef]
- Ikemura, T. Codon Usage and tRNA Content in Unicellular and Multicellular Organisms. Mol. Biol. Evol. 1985, 2, 13–34. [Google Scholar] [CrossRef]
- Shields, D.C.; Sharp, P.M.; Higgins, D.G.; Wright, F. “Silent” sites in Drosophila genes are not neutral: Evidence of selection among synonymous codons. Mol. Biol. Evol. 1988, 5, 704–716. [Google Scholar] [CrossRef] [Green Version]
- Poh, Y.-P.; Ting, C.-T.; Fu, H.-W.; Langley, C.H.; Begun, D.J. Population Genomic Analysis of Base Composition Evolution in Drosophila melanogaster. Genome Biol. Evol. 2012, 4, 1245–1255. [Google Scholar] [CrossRef] [Green Version]
- Lawrie, D.S.; Messer, P.W.; Hershberg, R.; Petrov, D.A. Strong Purifying Selection at Synonymous Sites in D. melanogaster. PLoS Genet. 2013, 9, e1003527. [Google Scholar] [CrossRef] [Green Version]
- Vicario, S.; Moriyama, E.N.; Powell, J.R. Codon usage in twelve species of Drosophila. BMC Evol. Biol. 2007, 7, 226. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, E.N.; Hartl, D.L. Codon usage bias and base composition of nuclear genes in Drosophila. Genetics 1993, 134, 847–858. [Google Scholar] [CrossRef]
- Powell, J.R.; Moriyama, E.N. Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. USA 1997, 94, 7784–7790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akashi, H. Inferring weak selection from patterns of polymorphism and divergence at ‘silent’ sites in Drosophila DNA. Genetics 1995, 139, 1067–1076. [Google Scholar] [CrossRef]
- Li, W.H. Models of nearly neutral mutations with particular implications for nonrandom usage of synonymous codons. J. Mol. Evol. 1987, 24, 337–345. [Google Scholar] [CrossRef]
- Kliman, R.M.; Hey, J. The effects of mutation and natural selection on codon bias in the genes of drosophila. Genetics 1994, 137, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Akashi, H. Translational selection and yeast proteome evolution. Genetics 2003, 164, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.L.; Moriyama, E.N.; Sawyer, S.A. Selection intensity for codon bias. Genetics 1994, 138, 227–234. [Google Scholar] [CrossRef]
- Ohta, T. Population size and rate of evolution. J. Mol. Evol. 1972, 1, 305–314. [Google Scholar] [CrossRef]
- Ohta, T. The Nearly Neutral Theory of Molecular Evolution. Annu. Rev. Ecol. Syst. 2003, 23, 263–286. [Google Scholar] [CrossRef]
- Matsuo, Y. Molecular evolution of the histone 3 multigene family in the Drosophila melanogaster species subgroup. Mol. Phylogenet. Evol. 2000, 16, 339–343. [Google Scholar] [CrossRef]
- Matsuo, Y. Evolutionary change of codon usage for the histone gene family in Drosophila melanogaster and Drosophila hydei. Mol. Phylogenet. Evol. 2000, 15, 283–291. [Google Scholar] [CrossRef]
- Matsuo, Y. Evolution of the GC content of the histone 3 gene in seven Drosophila species. Genes Genet. Syst. 2003, 78, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, Y.; Higashiyama, A.; Ushimaru, A.; Nagoda, N.; Matsuo, Y. Evolution of GC content in the histone gene repeatingunits from Drosophila lutescens, D. takahashii and D. pseudoobscura. Genes Genet. Syst. 2016, 91, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Kokko, H.; Eyre-Walker, A. Population size and the rate of evolution. Trends Ecol. Evol. 2014, 29, 33–41. [Google Scholar] [CrossRef]
- Bernardi, G.; Bernardi, G. Compositional constraints and genome evolution. J. Mol. Evol. 1986, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fitch, D.H.; Strausbaugh, L.D. Low codon bias and high rates of synonymous substitution in Drosophila hydei and D. melanogaster histone genes. Mol. Biol. Evol. 1993, 10, 397–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Trelles, F.; Tarrío, R.; Ayala, F.J. Switch in codon bias and increased rates of amino acid substitution in the Drosophila saltans species group. Genetics 1999, 153, 339–350. [Google Scholar] [CrossRef]
- Rodríguez-Trelles, F.; Tarrío, R.; Ayala, F.J. Fluctuating mutation bias and the evolution of base composition in Drosophila. J. Mol. Evol. 2000, 50, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Tsunemoto, K.; Matsuo, Y.; Tsunemotoa, K.; Matsuo, Y.; Tsunemoto, K.; Matsuo, Y. Molecular evolutionary analysis of a histone gene repeating unit from Drosophila simulans. Genes Genet. Syst. 2001, 76, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Nagoda, N.; Fukuda, A.; Nakashima, Y.; Matsuo, Y. Molecular characterization and evolution of the repeating units of histone genes in Drosophila americana: Coexistence of quartet and quintet units in a genome. Insect Mol. Biol. 2005, 14, 713–717. [Google Scholar] [CrossRef]
- Kakita, M.; Shimizu, T.; Emoto, M.; Nagai, M.; Takeguchi, M.; Hosono, Y.; Kume, N.; Ozawa, T.; Ueda, M.; Bhuiyan, M.S.I.; et al. Divergence and heterogeneity of the histone gene repeating units in the Drosophila melanogaster species subgroup. Genes Genet. Syst. 2003, 78, 383–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y. Epigenetics and Codon Usage of the Histone Genes in 12 Drosophila Species. J. Phylogenet. Evol. Biol. 2017, 5, 1–7. [Google Scholar] [CrossRef]
- Matsuo, Y.; Kakubayashi, N. Epigenetics Evolution and Replacement Histones: Evolutionary Changes at Drosophila H3.3A and H3.3B. J. Phylogenet. Evol. Biol. 2016, 4, 1–8. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Watanabe, T.; Nakamura, M.; Kakubayashi, N.; Saito, Y.; Matsuo, Y. Epigenetics Evolution and Replacement Histones: Evolutionary Changes at Drosophila H4r. J. Phylogenet. Evol. Biol. 2016, 4, 3. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, Y.; Kakubayashi, N. Epigenetics Evolution and Replacement Histones: Evolutionary Changes at Drosophila H2AvD. J. Data Min. Genom. Proteom. 2017, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Mukai, T. The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 1964, 50, 1–19. [Google Scholar] [CrossRef]
- Eyre-Walker, A.; Keightley, P.D. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 2007, 8, 610–618. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuo, Y. The Adenine/Thymine Deleterious Selection Model for GC Content Evolution at the Third Codon Position of the Histone Genes in Drosophila. Genes 2021, 12, 721. https://doi.org/10.3390/genes12050721
Matsuo Y. The Adenine/Thymine Deleterious Selection Model for GC Content Evolution at the Third Codon Position of the Histone Genes in Drosophila. Genes. 2021; 12(5):721. https://doi.org/10.3390/genes12050721
Chicago/Turabian StyleMatsuo, Yoshinori. 2021. "The Adenine/Thymine Deleterious Selection Model for GC Content Evolution at the Third Codon Position of the Histone Genes in Drosophila" Genes 12, no. 5: 721. https://doi.org/10.3390/genes12050721
APA StyleMatsuo, Y. (2021). The Adenine/Thymine Deleterious Selection Model for GC Content Evolution at the Third Codon Position of the Histone Genes in Drosophila. Genes, 12(5), 721. https://doi.org/10.3390/genes12050721




