Changes in Lactate Production, Lactate Dehydrogenase Genes Expression and DNA Methylation in Response to Tamoxifen Resistance Development in MCF-7 Cell Line
Abstract
:1. Introduction
2. Materials and Methods
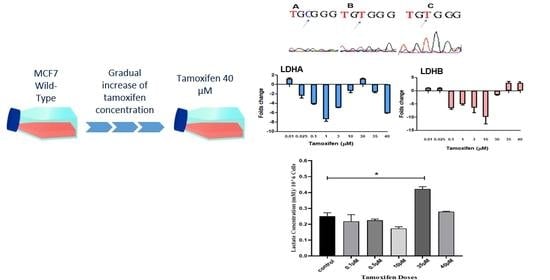
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anastasiadi, Z.; Lianos, G.; Ignatiadou, E.; Harissis, H.; Mitsis, M. Breast cancer in young women: An overview. Updates Surg. 2017, 69, 313–317. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Sauer, A.G.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Wang, Y.C.; Peto, R.; Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [PubMed] [Green Version]
- Lee, S.; Lee, H.; Jeong, D.; Ham, J.; Park, S.; Choi, E.; Kim, S. Cold atmospheric plasma restores tamoxifen sensitivity in resistant MCF-7 breast cancer cell. Free Radic. Biol. Med. 2017, 110, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Rondón-Lagos, M.; Villegas, V.E.; Rangel, N.; Sánchez, M.C.; Zaphiropoulos, P.G. Tamoxifen resistance: Emerging molecular targets. Int. J. Mol. Sci. 2016, 17, 1357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhong, Q.; Rhodes, L.; Townley, I.; Bratton, M.; Zhang, Q.; Martin, E.; Elliott, S.; Collins-Burow, B.; Burow, M.; et al. Proteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migration. Breast Cancer Res. 2012, 14, R45. [Google Scholar] [CrossRef] [Green Version]
- Shibata, T.; Watari, K.; Izumi, H.; Kawahara, A.; Hattori, S.; Fukumitsu, C.; Murakami, Y.; Takahashi, R.; Toh, U.; Ito, K.; et al. Breast Cancer Resistance to Antiestrogens Is Enhanced by Increased ER Degradation and ERBB2 Expression. Cancer Res. 2016, 77, 545–556. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Li, R.; Shi, W.; Jiang, T.; Wang, Y.; Li, C.; Qu, X. Silencing of MicroRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed. Pharmacother. 2016, 77, 37–44. [Google Scholar] [CrossRef]
- Tokunaga, E.; Kimura, Y.; Mashino, K.; Oki, E.; Kataoka, A.; Ohno, S.; Morita, M.; Kakeji, Y.; Baba, H.; Maehara, Y. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer 2006, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Huang, X.; Ye, F.; Chen, B.; Song, C.; Wen, J.; Zhang, Z.; Zheng, G.; Tang, H.; Xie, X. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci. Rep. 2016, 6, 21735. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.; Higham, S.; Perunovic, B.; Arafa, M.; Balasubramanian, S.; Rehman, I. Lactate Dehydrogenase-B Is Silenced by Promoter Methylation in a High Frequency of Human Breast Cancers. PLoS ONE 2013, 8, e57697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCleland, M.; Adler, A.; Shang, Y.; Hunsaker, T.; Truong, T.; Peterson, D.; Torres, E.; Li, L.; Haley, B.; Stephan, J.; et al. An Integrated Genomic Screen Identifies LDHB as an Essential Gene for Triple-Negative Breast Cancer. Cancer Res. 2012, 72, 5812–5823. [Google Scholar] [CrossRef] [Green Version]
- Leiblich, A.; Cross, S.S.; Catto, J.W.; Phillips, J.T.; Leung, H.Y.; Hamdy, F.C.; Rehman, I. Lactate dehydrogenase-B is silenced by promoter hypermethylation in human prostate cancer. Oncogene 2006, 25, 2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maekawa, M.; Taniguchi, T.; Ishikawa, J.; Sugimura, H.; Sugano, K.; Kanno, T. Promoter hypermethylation in cancer silences LDHB, eliminating lactate dehydrogenase isoenzymes 1–4. Clin. Chem. 2003, 49, 1518–1520. [Google Scholar] [CrossRef]
- Alhusban, A.A.; Breadmore, M.C.; Gueven, N.; Guijt, R.M. Capillary electrophoresis for automated on-line monitoring of suspension cultures: Correlating cell density, nutrients and metabolites in near real-time. Anal. Chim. Acta 2016, 920, 94–101. [Google Scholar] [CrossRef]
- Alhusban, A.A.; Breadmore, M.C.; Gueven, N.; Guijt, R.M. Time-Resolved Pharmacological Studies using Automated, On-line Monitoring of Five Parallel Suspension Cultures. Sci. Rep. 2017, 7, 1–9. [Google Scholar]
- Hamadneh, L.; Abuarqoub, R.; Alhusban, A.; Bahader, M. Upregulation of PI3K/AKT/PTEN pathway is correlated with glucose and glutamine metabolic dysfunction during tamoxifen resistance development in MCF-7 cells. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Mishra, D.; Banerjee, D. Lactate Dehydrogenases as Metabolic Links between Tumor and Stroma in the Tumor Microenvironment. Cancers 2019, 11, 750. [Google Scholar] [CrossRef] [Green Version]
- San-Millán, I.; Julian, C.G.; Matarazzo, C.; Martinez, J.; Brooks, G.A. Is lactate an oncometabolite? Evidence supporting a role for lactate in the regulation of transcriptional activity of cancer-related genes in MCF7 breast cancer cells. Front. Oncol. 2020, 9, 1536. [Google Scholar] [CrossRef] [PubMed]
- Billiard, J.; Dennison, J.B.; Briand, J.; Annan, R.S.; Chai, D.; Colón, M.; Dodson, C.S.; Gilbert, S.A.; Greshock, J.; Jing, J.; et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Zhao, Y.; Ding, Y.; Liu, H.; Liu, Z.; Fodstad, O.; Riker, A.I.; Kamarajugadda, S.; Lu, J.; Owen, L.B.; et al. Warburg effect in chemosensitivity: Targeting lactate dehydrogenase-A re-sensitizes Taxol-resistant cancer cells to Taxol. Mol. Cancer 2010, 9, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, Y.S.; Huang, Y.F.; Midha, M.K.; Chen, T.H.; Shiau, H.C.; Chiu, K.P. Single cell transcriptome analysis of MCF-7 reveals consistently and inconsistently expressed gene groups each associated with distinct cellular localization and functions. PLoS ONE 2018, 13, e0199471. [Google Scholar]
- Kondaveeti, Y.; Reed, I.K.; White, B.A. Epithelial–mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett. 2015, 364, 44–58. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- De la Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front. Oncol. 2019, 9, 1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamadneh, L.; Al-Lakkis, L.; Alhusban, A.A.; Tarawneh, S.; Abu-Irmaileh, B.; Albustanji, S.; Al-Bawab, A.Q. Changes in Lactate Production, Lactate Dehydrogenase Genes Expression and DNA Methylation in Response to Tamoxifen Resistance Development in MCF-7 Cell Line. Genes 2021, 12, 777. https://doi.org/10.3390/genes12050777
Hamadneh L, Al-Lakkis L, Alhusban AA, Tarawneh S, Abu-Irmaileh B, Albustanji S, Al-Bawab AQ. Changes in Lactate Production, Lactate Dehydrogenase Genes Expression and DNA Methylation in Response to Tamoxifen Resistance Development in MCF-7 Cell Line. Genes. 2021; 12(5):777. https://doi.org/10.3390/genes12050777
Chicago/Turabian StyleHamadneh, Lama, Lara Al-Lakkis, Ala A. Alhusban, Shahd Tarawneh, Bashaer Abu-Irmaileh, Sokiyna Albustanji, and Abdel Qader Al-Bawab. 2021. "Changes in Lactate Production, Lactate Dehydrogenase Genes Expression and DNA Methylation in Response to Tamoxifen Resistance Development in MCF-7 Cell Line" Genes 12, no. 5: 777. https://doi.org/10.3390/genes12050777
APA StyleHamadneh, L., Al-Lakkis, L., Alhusban, A. A., Tarawneh, S., Abu-Irmaileh, B., Albustanji, S., & Al-Bawab, A. Q. (2021). Changes in Lactate Production, Lactate Dehydrogenase Genes Expression and DNA Methylation in Response to Tamoxifen Resistance Development in MCF-7 Cell Line. Genes, 12(5), 777. https://doi.org/10.3390/genes12050777