PPARG Hypermethylation as the First Epigenetic Modification in Newly Onset Insulin Resistance in Human Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Preadipocyte Collection and Differentiation
2.2. T3-L1 Cell Line Culture and Differentiation
2.3. Insulin Resistance Induction
2.4. Genetic Material Extraction
2.5. Gene Expression
2.6. Global and Site-Specific DNA Methylation
2.7. Statistical Analysis
3. Results
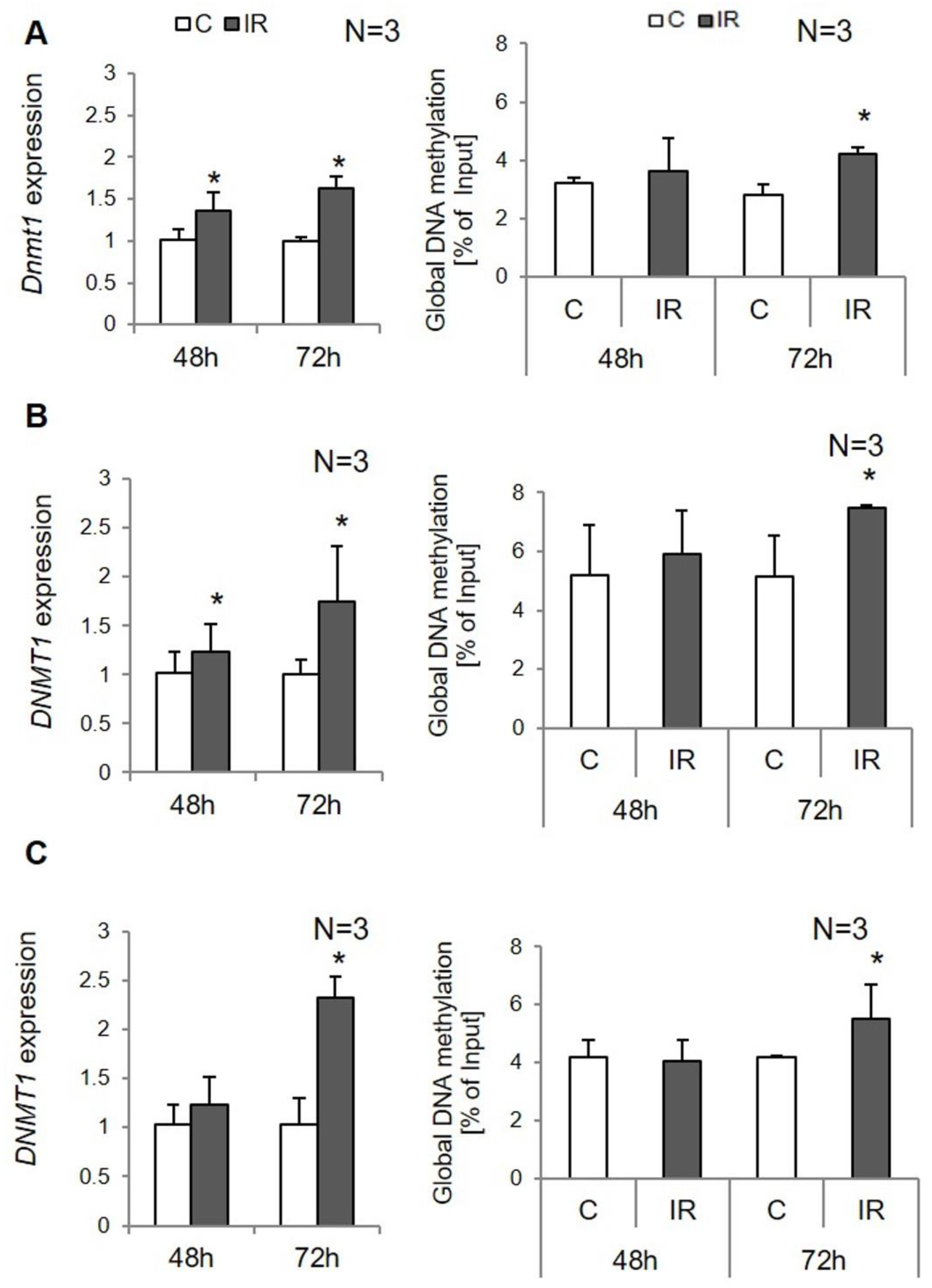
3.1. Global DNA Methylation in Insulin-Resistant Adipocytes
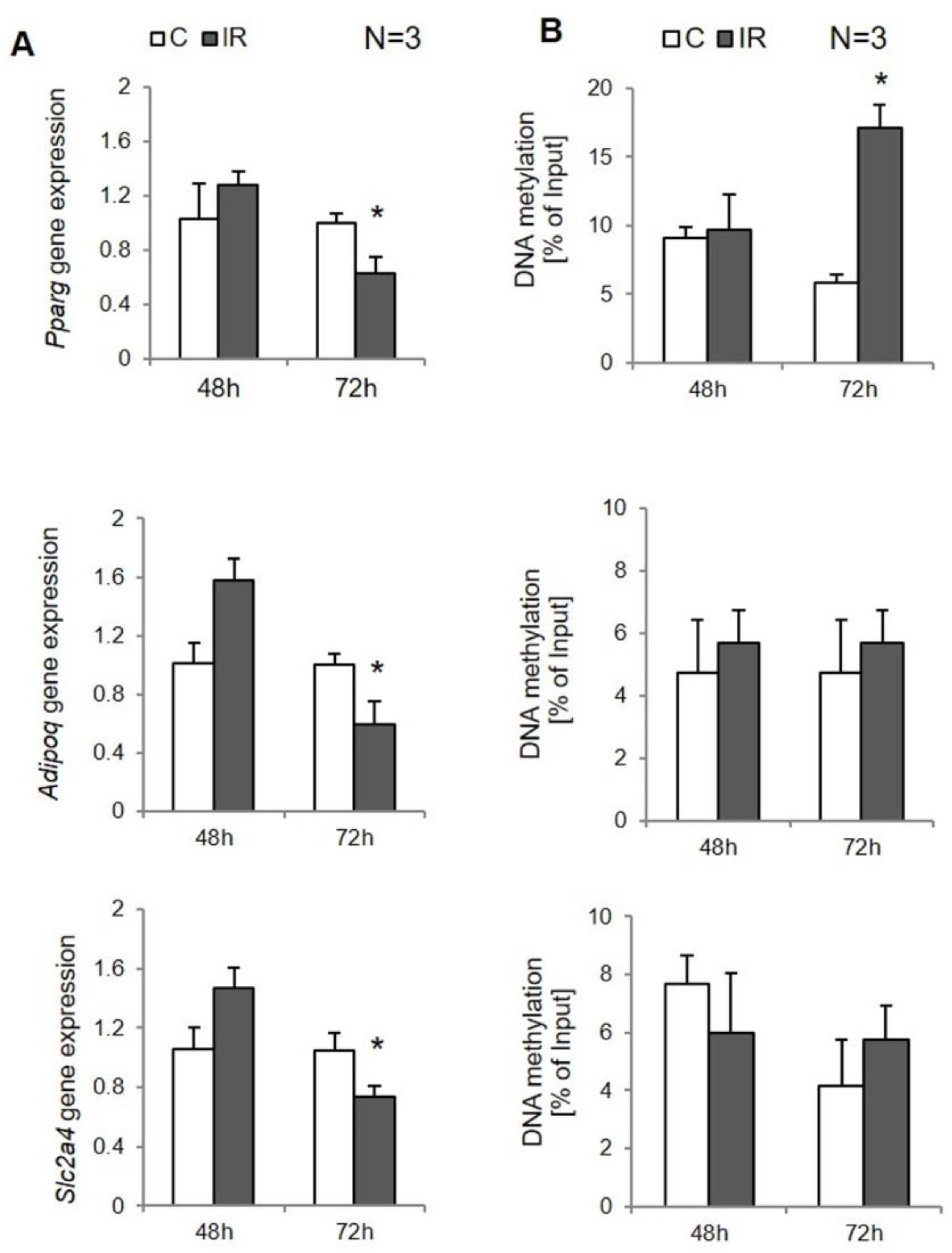
3.2. Site-Specific DNA Methylation in Insulin-Resistant 3T3-L1 Adipocytes
3.3. Site-Specific DNA Methylation in Insulin-Resistant Human Adipocytes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lebovitz, H.E. Insulin Resistance: Definition and Consequences. Exp. Clin. Endocrinol. Diabetes 2001, 109 (Suppl. 2), S135–S148. [Google Scholar] [CrossRef] [Green Version]
- Abdul-Ghani, M.A.; DeFronzo, R.A. Pathogenesis of Insulin Resistance in Skeletal Muscle. J. Biomed. Biotechnol. 2010, 2010, 476279. [Google Scholar] [CrossRef] [Green Version]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [Green Version]
- Engin, A. The Definition and Prevalence of Obesity and Metabolic Syndrome. In Obesity and Lipotoxicity; Engin, A.B., Engin, A., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switerland, 2017; pp. 1–17. ISBN 978-3-319-48382-5. [Google Scholar]
- Semple, R.K. PPAR and Human Metabolic Disease. J. Clin. Investig. 2006, 116, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Malodobra-Mazur, M.; Cierzniak, A.; Dobosz, T. Oleic Acid Influences the Adipogenesis of 3T3-L1 Cells via DNA Methylation and May Predispose to Obesity and Obesity-Related Disorders. Lipids Health Dis. 2019, 18, 230. [Google Scholar] [CrossRef] [Green Version]
- Motojima, K.; Passilly, P.; Peters, J.M.; Gonzalez, F.J.; Latruffe, N. Expression of Putative Fatty Acid Transporter Genes Are Regulated by Peroxisome Proliferator-Activated Receptor Alpha and Gamma Activators in a Tissue- and Inducer-Specific Manner. J. Biol. Chem. 1998, 273, 16710–16714. [Google Scholar] [CrossRef] [Green Version]
- Sironi, A.M.; Vichi, S.; Gastaldelli, A.; Pecori, N.; Anichini, R.; Foot, E.; Seghieri, G.; Ferrannini, E. Effects of Troglitazone on Insulin Action and Cardiovascular Risk Factors in Patients with Non-Insulin-Dependent Diabetes. Clin. Pharm. Ther. 1997, 62, 194–202. [Google Scholar] [CrossRef]
- González-Becerra, K.; Ramos-Lopez, O.; Barrón-Cabrera, E.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez-López, E.; Martínez, J.A. Fatty Acids, Epigenetic Mechanisms and Chronic Diseases: A Systematic Review. Lipids Health Dis. 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z. The Metabolic Syndrome. Lancet 2005, 365, 1415–1428. [Google Scholar] [CrossRef]
- Cierzniak, A.; Pawelka, D.; Kaliszewski, K.; Rudnicki, J.; Dobosz, T.; Malodobra-Mazur, M. DNA Methylation in Adipocytes from Visceral and Subcutaneous Adipose Tissue Influences Insulin-Signaling Gene Expression in Obese Individuals. Int. J. Obes. 2021, 45, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Chillarón, J.C.; Díaz, R.; Martínez, D.; Pentinat, T.; Ramón-Krauel, M.; Ribó, S.; Plösch, T. The Role of Nutrition on Epigenetic Modifications and Their Implications on Health. Biochimie 2012, 94, 2242–2263. [Google Scholar] [CrossRef]
- Deans, C.; Maggert, K.A. What Do You Mean, “Epigenetic”? Genetics 2015, 199, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Deaton, A.M.; Bird, A. CpG Islands and the Regulation of Transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Małodobra-Mazur, M.; Alama, A.; Bednarska-Chabowska, D.; Pawelka, D.; Myszczyszyn, A.; Dobosz, T. Obesity-Induced Insulin Resistance via Changes in the DNA Methylation Profile of Insulin Pathway Genes. Adv. Clin. Exp. Med. 2019, 28, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Goldberg, J.; Bremner, J.D.; Vaccarino, V. Global DNA Methylation Is Associated with Insulin Resistance: A Monozygotic Twin Study. Diabetes 2012, 61, 542–546. [Google Scholar] [CrossRef] [Green Version]
- Grundberg, E.; Meduri, E.; Sandling, J.K.; Hedman, A.K.; Keildson, S.; Buil, A.; Busche, S.; Yuan, W.; Nisbet, J.; Sekowska, M.; et al. Global Analysis of DNA Methylation Variation in Adipose Tissue from Twins Reveals Links to Disease-Associated Variants in Distal Regulatory Elements. Am. J. Hum. Genet. 2013, 93, 876–890. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo, B.; Irvin, M.R.; Sha, J.; Zhi, D.; Aslibekyan, S.; Absher, D.; Tiwari, H.K.; Kabagambe, E.K.; Ordovas, J.M.; Arnett, D.K. Epigenome-Wide Association Study of Fasting Measures of Glucose, Insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network Study. Diabetes 2014, 63, 801–807. [Google Scholar] [CrossRef] [Green Version]
- Małodobra-Mazur, M.; Cierzniak, A.; Pawełka, D.; Kaliszewski, K.; Rudnicki, J.; Dobosz, T. Metabolic Differences between Subcutaneous and Visceral Adipocytes Differentiated with an Excess of Saturated and Monounsaturated Fatty Acids. Genes 2020, 11, 1092. [Google Scholar] [CrossRef]
- Gosmain, Y.; Lefai, E.; Ryser, S.; Roques, M.; Vidal, H. Sterol Regulatory Element-Binding Protein-1 Mediates the Effect of Insulin on Hexokinase II Gene Expression in Human Muscle Cells. Diabetes 2004, 53, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Accili, D.; Arden, K.C. FoxOs at the Crossroads of Cellular Metabolism, Differentiation, and Transformation. Cell 2004, 117, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Pinel, A.; Rigaudière, J.-P.; Jouve, C.; Capel, F. Modulation of Insulin Resistance and the Adipocyte-Skeletal Muscle Cell Cross-Talk by LCn-3PUFA. Int. J. Mol. Sci. 2018, 19, 2778. [Google Scholar] [CrossRef] [Green Version]
- Shinjo, S.; Jiang, S.; Nameta, M.; Suzuki, T.; Kanai, M.; Nomura, Y.; Goda, N. Disruption of the Mitochondria-Associated ER Membrane (MAM) Plays a Central Role in Palmitic Acid-Induced Insulin Resistance. Exp. Cell Res. 2017, 359, 86–93. [Google Scholar] [CrossRef]
- Kim, A.Y.; Park, Y.J.; Pan, X.; Shin, K.C.; Kwak, S.-H.; Bassas, A.F.; Sallam, R.M.; Park, K.S.; Alfadda, A.A.; Xu, A.; et al. Obesity-Induced DNA Hypermethylation of the Adiponectin Gene Mediates Insulin Resistance. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.H.; An, S.M.; Ye, B.J.; Lee, J.H.; Yoo, E.J.; Jeong, G.W.; Kang, H.J.; Alfadda, A.A.; Lim, S.W.; Park, J.; et al. TonEBP/NFAT5 Promotes Obesity and Insulin Resistance by Epigenetic Suppression of White Adipose Tissue Beiging. Nat. Commun. 2019, 10, 3536. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cao, Q.; Yu, L.; Shi, H.; Xue, B.; Shi, H. Epigenetic Regulation of Macrophage Polarization and Inflammation by DNA Methylation in Obesity. JCI Insight 2016, 1, e87748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, E.; Jansson, P.A.; Perfilyev, A.; Volkov, P.; Pedersen, M.; Svensson, M.K.; Poulsen, P.; Ribel-Madsen, R.; Pedersen, N.L.; Almgren, P.; et al. Altered DNA Methylation and Differential Expression of Genes Influencing Metabolism and Inflammation in Adipose Tissue from Subjects with Type 2 Diabetes. Diabetes 2014, 63, 2962–2976. [Google Scholar] [CrossRef] [Green Version]
- Esteves, J.V.; Yonamine, C.Y.; Machado, U.F. SLC2A4 Expression and Its Epigenetic Regulation as Biomarkers for Insulin Resistance Treatment in Diabetes Mellitus. Biomark. Med. 2020, 14, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Kubota, N.; Terauchi, Y.; Miki, H.; Tamemoto, H.; Yamauchi, T.; Komeda, K.; Satoh, S.; Nakano, R.; Ishii, C.; Sugiyama, T.; et al. PPAR Gamma Mediates High-Fat Diet-Induced Adipocyte Hypertrophy and Insulin Resistance. Mol. Cell 1999, 4, 597–609. [Google Scholar] [CrossRef]
- Davé, V.; Yousefi, P.; Huen, K.; Volberg, V.; Holland, N. Relationship between Expression and Methylation of Obesity-Related Genes in Children. Mutagenesis 2015, 30, 411–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | Species | Sequence [3′→5′] | Amplicon [bp] | CpG * | |
|---|---|---|---|---|---|
| PPARG | Human | F | CTGTTATGGGTGAAACTCTGG | 58 | 4 |
| R | GTGAAGGAATCGCTTTCTGG | ||||
| SLC2A4 | Human | F | TTGTGGCTGTGGGTCCCAT | 153 | 15 |
| R | CTCGTCTTAGAAGAGCTGGA | ||||
| ADIPOQ | Human | F | GCTGTTCTACTGCTATTAGC | 196 | 6 |
| R | GATCTCCTTTCTCACCCTTC | ||||
| Pparg | Mouse | F | ACACCAGTGTGAATTACAGC | 79 | 2 |
| R | TCTGGGTCAACAGGAGAAATC | ||||
| Slc2a4 | Mouse | F | CAAGCGGGTCTCACTAGATC | 176 | 13 |
| R | AGACTCAGGCGCTGCAATAA | ||||
| Adipoq | Mouse | F | CCTGTTCCTCTTAATCCTGC | 96 | 3 |
| R | CAAGTTCCCTTGGGTGGAG | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Małodobra-Mazur, M.; Cierzniak, A.; Kaliszewski, K.; Dobosz, T. PPARG Hypermethylation as the First Epigenetic Modification in Newly Onset Insulin Resistance in Human Adipocytes. Genes 2021, 12, 889. https://doi.org/10.3390/genes12060889
Małodobra-Mazur M, Cierzniak A, Kaliszewski K, Dobosz T. PPARG Hypermethylation as the First Epigenetic Modification in Newly Onset Insulin Resistance in Human Adipocytes. Genes. 2021; 12(6):889. https://doi.org/10.3390/genes12060889
Chicago/Turabian StyleMałodobra-Mazur, Małgorzata, Aneta Cierzniak, Krzysztof Kaliszewski, and Tadeusz Dobosz. 2021. "PPARG Hypermethylation as the First Epigenetic Modification in Newly Onset Insulin Resistance in Human Adipocytes" Genes 12, no. 6: 889. https://doi.org/10.3390/genes12060889








