Co-Regulated Genes and Gene Clusters
Abstract
:1. Introduction
2. Erythroid-Specific Genes
3. Functional Role of Gene Clustering in Vertebrate Cells
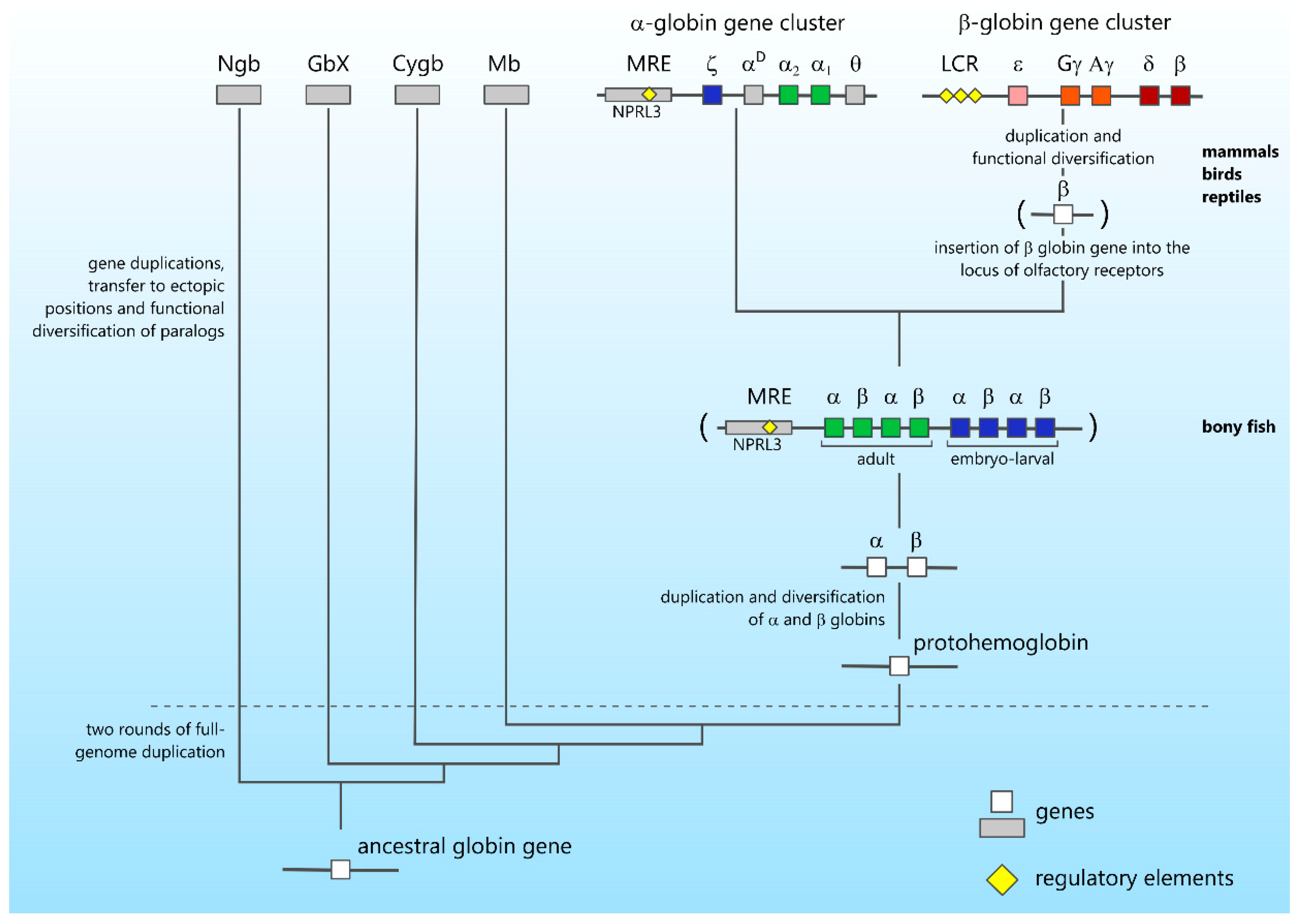
4. Gene Cluster Evolution
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.; Feng, J.; Yu, B.; Ma, Q.; Liu, B. The functional determinants in the organization of bacterial genomes. Brief. Bioinform. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Reznikoff, W.S. The Operon Revisited. Annu. Rev. Genet. 1972, 6, 133–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Blumenthal, T. Operon and non-operon gene clusters in the C. elegans genome. WormBook 2014, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blumenthal, T.; Evans, N.; Link, C.D.; Guffanti, A.; Lawson, D.; Thierry-Mieg, J.; Thierry-Mieg, D.; Chiu, W.L.; Duke, K.; Kiraly, M.; et al. A global analysis of Caenorhabditis elegans operons. Nature 2002, 417, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.L.; Blumenthal, T. Trans-splicing. Wiley Interdiscip. Rev. RNA 2011, 2, 417–434. [Google Scholar] [CrossRef]
- Wong, S.; Wolfe, K.H. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 2005, 37, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Hittinger, C.T.; Rokas, A.; Carroll, S.B. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc. Natl. Acad. Sci. USA 2004, 101, 14144–14149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Genet. 2012, 11, 21–32. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism from biochemistry to genomics. Nat. Rev. Genet. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.; Chomet, P.; Glawischnig, E.; Stettner, C.; Grün, S.; Winklmair, A.; Eisenreich, W.; Bacher, A.; Meeley, R.B.; Briggs, S.P.; et al. Analysis of a Chemical Plant Defense Mechanism in Grasses. Science 1997, 277, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, N.; Zhang, J.; Oyagbenro, R.K.; Brown, B.; Wu, Y.; Yang, B.; Li, Z.; Peters, R.J. Interdependent evolution of biosynthetic gene clusters for momilactone production in rice. Plant Cell 2021, 33, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Kawaide, H.; Higuchi, T.; Chen, M.; Miyamoto, K.; Hirata, Y.; Kimura, H.; Miyazaki, S.; Teruya, M.; Fujiwara, K.; et al. Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc. Natl. Acad. Sci. USA 2020, 117, 12472–12480. [Google Scholar] [CrossRef]
- Boycheva, S.; Daviet, L.; Wolfender, J.-L.; Fitzpatrick, T.B. The rise of operon-like gene clusters in plants. Trends Plant Sci. 2014, 19, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Collemare, J.; Seidl, M.F. Chromatin-dependent regulation of secondary metabolite biosynthesis in fungi: Is the picture complete? FEMS Microbiol. Rev. 2019, 43, 591–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurst, L.D.; Pál, C.; Lercher, M.J. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004, 5, 299–310. [Google Scholar] [CrossRef]
- Nützmann, H.-W.; Scazzocchio, C.; Osbourn, A. Metabolic Gene Clusters in Eukaryotes. Annu. Rev. Genet. 2018, 52, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Philipsen, S.; Hardison, R.C. Evolution of hemoglobin loci and their regulatory elements. Blood Cells Mol. Dis. 2018, 70, 2–12. [Google Scholar] [CrossRef]
- Ganis, J.J.; Hsia, N.; Trompouki, E.; De Jong, J.L.; DiBiase, A.; Lambert, J.S.; Jia, Z.; Sabo, P.J.; Weaver, M.; Sandstrom, R.; et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol. 2012, 366, 185–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardison, R.C. Evolution of Hemoglobin and Its Genes. Cold Spring Harb. Perspect. Med. 2012, 2, a011627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, C.; Burmester, T.; Hankeln, T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet. Genome Res. 2006, 112, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Recillas-Targa, F. Chromatin domains and regulation of gene expression: Familiar and enigmatic clusters of chicken globin genes. Crit. Rev. Eukaryot. Gene Expr. 2001, 11, 16. [Google Scholar] [CrossRef]
- Razin, S.V.; Farrell, C.M.; Recillas-Targa, F. Genomic Domains and Regulatory Elements Operating at the Domain Level. Int. Rev. Cytol. 2003, 226, 63–125. [Google Scholar] [CrossRef] [PubMed]
- Grosveld, F.; van Assandelt, G.B.; Greaves, D.R.; Kollias, B. Position-independent, high-level expression of the human b-globin gene in transgenic mice. Cell 1987, 51, 975–985. [Google Scholar] [CrossRef] [Green Version]
- Jarman, A.P.; Wood, W.G.; Sharpe, J.A.; Gourdon, G.; Ayyub, H.; Higgs, D.R. Characterization of the major regulatory element upstream of the human a-globin gene cluster. Mol. Cell. Biol. 1991, 11, 4679–4689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oudelaar, A.M.; A Beagrie, R.; Kassouf, M.T.; Higgs, D.R. The mouse alpha-globin cluster: A paradigm for studying genome regulation and organization. Curr. Opin. Genet. Dev. 2021, 67, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Tolhuis, B.; Palstra, R.-J.; Splinter, E.; Grosveld, F.; de Laat, W. Looping and Interaction between Hypersensitive Sites in the Active β-globin Locus. Mol. Cell 2002, 10, 1453–1465. [Google Scholar] [CrossRef]
- Vernimmen, D.; De Gobbi, M.; A Sloane-Stanley, J.; Wood, W.G.; Higgs, D.R. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007, 26, 2041–2051. [Google Scholar] [CrossRef]
- Vernimmen, D.; Marques-Kranc, F.; Sharpe, J.A.; Sloane-Stanley, J.A.; Wood, W.G.; Wallace, H.A.C.; Smith, A.J.H.; Higgs, D.R. Chromosome looping at the human α-globin locus is mediated via the major upstream regulatory element (HS −40). Blood 2009, 114, 4253–4260. [Google Scholar] [CrossRef] [Green Version]
- De Laat, W.; Grosveld, F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003, 11, 447–459. [Google Scholar] [CrossRef]
- De Laat, W.; Klous, P.; Kooren, J.; Noordermeer, D.; Palstra, R.J.; Simonis, M.; Splinter, E.; Grosveld, F. Three-dimensional organization of gene expression in erythroid cells. Curr. Top. Dev. Biol. 2008, 82, 117–139. [Google Scholar] [PubMed]
- Cantor, A.B.; Orkin, S.H. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene 2002, 21, 3368–3376. [Google Scholar] [CrossRef] [Green Version]
- Gurumurthy, A.; Shen, Y.; Gunn, E.M.; Bungert, J. Phase Separation and Transcription Regulation: Are Super-Enhancers and Locus Control Regions Primary Sites of Transcription Complex Assembly? BioEssays 2018, 41, e1800164. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cairns, M.J.; Yan, J. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 2019, 47, 11481–11496. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Song, S.-H.; Lee, J.J.; Choi, N.; Kim, C.G.; Dean, A.; Kim, A. The role of transcriptional activator GATA-1 at human -globin HS2. Nucleic Acids Res. 2008, 36, 4521–4528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, H.; Grass, J.A.; Johnson, K.D.; Kim, S.-I.; Boyer, M.E.; Imbalzano, A.N.; Bieker, J.J.; Bresnick, E.H. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. USA 2005, 102, 17065–17070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawado, T.; Igarashi, K.; Groudine, M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 2001, 98, 10226–10231. [Google Scholar] [CrossRef] [Green Version]
- Gurumurthy, A.; Yu, D.T.; Stees, J.R.; Chamales, P.; Gavrilova, E.; Wassel, P.; Li, L.; Stribling, D.; Chen, J.; Brackett, M.; et al. Super-enhancer mediated regulation of adult β-globin gene expression: The role of eRNA and Integrator. Nucleic Acids Res. 2021, 49, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razin, S.V.; Gavrilov, A.A. The Role of Liquid–Liquid Phase Separation in the Compartmentalization of Cell Nucleus and Spatial Genome Organization. Biochemistry 2020, 85, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, K.; Johnson, K.; Gao, X.; Keles, S.; Bresnick, E. The Hematopoietic Stem and Progenitor Cell Cistrome. Curr. Top. Dev. Biol. 2016, 118, 45–76. [Google Scholar] [CrossRef] [PubMed]
- Craddock, C.; Vyas, P.; Sharpe, J.; Ayyub, H.; Wood, W.; Higgs, D. Contrasting effects of alpha and beta globin regulatory elements on chromatin structure may be related to their different chromosomal environments. EMBO J. 1995, 14, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, E.C.; Bresnick, E.H. Histone acetylation beyond promoters: Long-range acetylation patterns in the chromatin world. BioEssays 2001, 23, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, E.C.; Downs, K.M.; Christensen, H.M.; Im, H.; Nuzzi, P.A.; Bresnick, E.H. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 2000, 97, 14494–14499. [Google Scholar] [CrossRef] [Green Version]
- Forrester, W.C.; Epner, E.; Driscoll, M.C.; Enver, T.; Brice, M.; Papayannopoulou, T.; Groudine, M. A deletion of the human b-globin locus activation region causes a major alteration in chromatin structure and replication across the entire b-globin locus. Gene Dev. 1990, 4, 1637–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyas, P.; Vickers, M.; Simmons, D.; Ayyub, H.; Craddock, C.; Higgs, D. Cis-acting sequences regulating expression of the human α-globin cluster lie within constitutively open chromatin. Cell 1992, 69, 781–793. [Google Scholar] [CrossRef]
- Zhou, G.-L.; Xin, L.; Song, W.; Di, L.-J.; Liu, G.; Wu, X.-S.; Liu, D.-P.; Liang, C.-C. Active Chromatin Hub of the Mouse α-Globin Locus Forms in a Transcription Factory of Clustered Housekeeping Genes. Mol. Cell. Biol. 2006, 26, 5096–5105. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.S.; McColl, B.; Maksimovic, J.; Vadolas, J. Epigenetic interplay at the β-globin locus. Biochim. Biophys. Acta Bioenerg. 2017, 1860, 393–404. [Google Scholar] [CrossRef]
- Fromm, G.; Bulger, M. A spectrum of gene regulatory phenomena at mammalian β-globin gene lociThis paper is one of a selection of papers published in this Special Issue, entitled 30th Annual International Asilomar Chromatin and Chromosomes Conference, and has undergone the Journal’s usual peer review process. Biochem. Cell Biol. 2009, 87, 781–790. [Google Scholar] [CrossRef]
- Kim, Y.W.; Yun, W.J.; Kim, A. Erythroid activator NF-E2, TAL1 and KLF1 play roles in forming the LCR HSs in the human adult β-globin locus. Int. J. Biochem. Cell Biol. 2016, 75, 45–52. [Google Scholar] [CrossRef]
- Su, M.Y.; Steiner, L.A.; Bogardus, H.; Mishra, T.; Schulz, V.; Hardison, R.C.; Gallagher, P.G. Identification of Biologically Relevant Enhancers in Human Erythroid Cells*. J. Biol. Chem. 2013, 288, 8433–8444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Q.; Cheng, L.; Tang, X.; He, Y.; Li, Y.; Yee, T.; Shrestha, D.; Feng, R.; Xu, P.; Zhou, X.; et al. Dynamic CTCF binding directly mediates interactions among cis-regulatory elements essential for hematopoiesis. Blood 2021, 137, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Sawaengdee, W.; Cui, K.; Zhao, K.; Hongeng, S.; Fucharoen, S.; Wongtrakoongate, P. Genome-Wide Transcriptional Regulation of the Long Non-coding RNA Steroid Receptor RNA Activator in Human Erythroblasts. Front. Genet. 2020, 11, 850. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.; Sexton, T.; Chakalova, L.; Cope, N.F.; Horton, A.; Andrews, S.; Kurukuti, S.; A Mitchell, J.; Umlauf, D.; Dimitrova, D.S.; et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2009, 42, 53–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.M.; Leach, J.; Reittie, J.E.; Atzberger, A.; E Leeprudhoe, J.; Wood, W.G.; Higgs, D.R.; Iborra, F.J.; Buckle, V.J. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006, 172, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, A.A.; Razin, S.V. Spatial configuration of the chicken α-globin gene domain: Immature and active chromatin hubs. Nucleic Acids Res. 2008, 36, 4629–4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulianov, S.V.; A Gavrilov, A.; Razin, S.V. Spatial organization of the chicken beta-globin gene domain in erythroid cells of embryonic and adult lineages. Epigenetics Chromatin 2012, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Kolodziej, K.E.; Obara, N.; Amaral-Psarris, A.; Demmers, J.; Shi, L.; Engel, J.D.; Grosveld, F.; Strouboulis, J.; Tanabe, O. Nuclear Receptors TR2 and TR4 Recruit Multiple Epigenetic Transcriptional Corepressors That Associate Specifically with the Embryonic β-Type Globin Promoters in Differentiated Adult Erythroid Cells. Mol. Cell. Biol. 2011, 31, 3298–3311. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Plank-Bazinet, J.; Krivega, I.; Dale, R.K.; Dean, A. Embryonic erythropoiesis and hemoglobin switching require transcriptional repressor ETO2 to modulate chromatin organization. Nucleic Acids Res. 2020, 48, 10226–10240. [Google Scholar] [CrossRef]
- Liu, N.; Hargreaves, V.V.; Zhu, Q.; Kurland, J.V.; Hong, J.; Kim, W.; Sher, F.; Trevino, C.M.; Rogers, J.M.; Kurita, R.; et al. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 2018, 173, 430–442.e17. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Jearawiriyapaisarn, N.; Lee, M.P.; Hosoya, T.; Wu, Q.; Myers, G.; Lim, K.-C.; Kurita, R.; Nakamura, Y.; Vojtek, A.B.; et al. BAP1 regulation of the key adaptor protein NCoR1 is critical for γ-globin gene repression. Genes Dev. 2018, 32, 1537–1549. [Google Scholar] [CrossRef] [Green Version]
- Iarovaia, O.V.; Kovina, A.P.; Petrova, N.; Razin, S.V.; Ioudinkova, E.S.; Vassetzky, Y.; Ulianov, S.V. Genetic and Epigenetic Mechanisms of β-Globin Gene Switching. Biochemistry 2018, 83, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Galitsyna, A.A.; Flyamer, I.M.; Golov, A.; Khrameeva, E.E.; Imakaev, M.V.; Abdennur, N.A.; Gelfand, M.S.; Gavrilov, A.A.; Razin, S.V. Activation of the alpha-globin gene expression correlates with dramatic upregulation of nearby non-globin genes and changes in local and large-scale chromatin spatial structure. Epigenetics Chromatin 2017, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanssen, L.; Kassouf, M.T.; Oudelaar, A.M.; Biggs, D.; Preece, C.; Downes, D.J.; Gosden, M.; Sharpe, J.A.; Sloane-Stanley, J.A.; Hughes, J.R.; et al. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017, 19, 952–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.W.; Kang, Y.; Kang, J.; Kim, A. GATA-1-dependent histone H3K27 acetylation mediates erythroid cell-specific chromatin interaction between CTCF sites. FASEB J. 2020, 34, 14736–14749. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Kim, Y.W.; Kang, J.; Yun, W.J.; Kim, A. Erythroid specific activator GATA-1-dependent interactions between CTCF sites around the β-globin locus. Biochim. Biophys. Acta Bioenerg. 2017, 1860, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, Z.; Huang, Y.; Liu, D.-P.; Liu, G.; Shen, W.; Tang, X.; Feng, D.; Liang, C.-C. Gene order in human α-globin locus is required for their temporal specific expressions. Genes Cells 2005, 11, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Harju, S.; Navas, P.A.; Stamatoyannopoulos, G.; Peterson, K.R. Genome Architecture of the Human β-Globin Locus Affects Developmental Regulation of Gene Expression. Mol. Cell. Biol. 2005, 25, 8765–8778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanimoto, K.; Liu, Q.; Bungert, J.; Engel, J.D. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature 1999, 398, 344–348. [Google Scholar] [CrossRef]
- A Lloyd, J.; Krakowsky, J.M.; Crable, S.C.; Lingrel, J.B. Human gamma- to beta-globin gene switching using a mini construct in transgenic mice. Mol. Cell. Biol. 1992, 12, 1561–1567. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.I.; Fiering, S.; Groudine, M. Regulation of β-globin gene expression: Straightening out the locus. Curr. Opin. Genet. Dev. 1996, 6, 488–495. [Google Scholar] [CrossRef]
- Soshnikova, N.; Duboule, D. Epigenetic Temporal Control of Mouse Hox Genes in Vivo. Science 2009, 324, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Montavon, T.; Soshnikova, N. Hox gene regulation and timing in embryogenesis. Semin. Cell Dev. Biol. 2014, 34, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, F.; Duboule, D.; Spitz, F. Transgenic analysis of Hoxd gene regulation during digit development. Dev. Biol. 2007, 306, 847–859. [Google Scholar] [CrossRef] [Green Version]
- Spitz, F.; Gonzalez, F.; Duboule, D. A Global Control Region Defines a Chromosomal Regulatory Landscape Containing the HoxD Cluster. Cell 2003, 113, 405–417. [Google Scholar] [CrossRef] [Green Version]
- Andrey, G.; Montavon, T.; Mascrez, B.; Gonzalez, F.; Noordermeer, D.; Leleu, M.; Trono, D.; Spitz, F.; Duboule, D. A Switch Between Topological Domains Underlies HoxD Genes Collinearity in Mouse Limbs. Science 2013, 340, 1234167. [Google Scholar] [CrossRef]
- Fabre, P.J.; Leleu, M.; Mormann, B.H.; Lopez-Delisle, L.; Noordermeer, D.; Beccari, L.; Duboule, D. Large scale genomic reorganization of topological domains at the HoxD locus. Genome Biol. 2017, 18, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Carballo, E.; Lopez-Delisle, L.; Willemin, A.; Beccari, L.; Gitto, S.; Mascrez, B.; Duboule, D. Chromatin topology and the timing of enhancer function at the HoxD locus. Proc. Natl. Acad. Sci. USA 2020, 117, 31231–31241. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carballo, E.; Lopez-Delisle, L.; Yakushiji-Kaminatsui, N.; Ullate-Agote, A.; Duboule, D. Impact of genome architecture on the functional activation and repression of Hox regulatory landscapes. BMC Biol. 2019, 17, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Vaes, E.; Mombaerts, P. Regulation of the Probability of Mouse Odorant Receptor Gene Choice. Cell 2011, 147, 907–921. [Google Scholar] [CrossRef] [Green Version]
- Fuss, S.H.; Omura, M.; Mombaerts, P. Local and cis Effects of the H Element on Expression of Odorant Receptor Genes in Mouse. Cell 2007, 130, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Serizawa, S.; Miyamichi, K.; Nakatani, H.; Suzuki, M.; Saito, M.; Yoshihara, Y.; Sakano, H. Negative Feedback Regulation Ensures the One Receptor-One Olfactory Neuron Rule in Mouse. Science 2003, 302, 2088–2094. [Google Scholar] [CrossRef] [Green Version]
- Monahan, K.; Horta, A.; Lomvardas, S. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nat. Cell Biol. 2019, 565, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Bashkirova, E.; Lomvardas, S. Olfactory receptor genes make the case for inter-chromosomal interactions. Curr. Opin. Genet. Dev. 2019, 55, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Niimura, Y.; Kobayashi, C.; Shirakawa, D.; Suzuki, H.; Enomoto, T.; Touhara, K.; Yoshihara, Y.; Hirota, J. A long-range cis-regulatory element for class I odorant receptor genes. Nat. Commun. 2017, 8, 885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.-C.; Cooke, N.E.; Liebhaber, S.A. Long-range looping of a locus control region drives tissue-specific chromatin packing within a multigene cluster. Nucleic Acids Res. 2016, 44, 4651–4664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Li, J.; Ge, X.; Wu, Y.; Guo, Y.; Wu, Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol. 2020, 21, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Monahan, K.; Wu, H.; Gertz, J.; Varley, K.; Li, W.; Myers, R.M.; Maniatis, T.; Wu, Q. CTCF/cohesin-mediated DNA looping is required for protocadherin promoter choice. Proc. Natl. Acad. Sci. USA 2012, 109, 21081–21086. [Google Scholar] [CrossRef] [Green Version]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffié, R.; Monahan, K.; O’Keeffe, S.; Simon, M.D.; et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 2019, 177, 639–653. [Google Scholar] [CrossRef] [Green Version]
- Almenar-Queralt, A.; Merkurjev, D.; Kim, H.S.; Navarro, M.; Ma, Q.; Chaves, R.S.; Allegue, C.; Driscoll, S.P.; Chen, A.G.; Kohlnhofer, B.; et al. Chromatin establishes an immature version of neuronal protocadherin selection during the naive-to-primed conversion of pluripotent stem cells. Nat. Genet. 2019, 51, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Haws, P.; Wu, Q. Multiple Variable First Exons: A Mechanism for Cell- and Tissue-Specific Gene Regulation. Genome Res. 2003, 14, 79–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hozumi, N.; Tonegawa, S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. 1976 [classical article]. J. Immunol. 2004, 173, 4260–4264. [Google Scholar] [PubMed]
- Michalak, P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 2008, 91, 243–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purmann, A.; Toedling, J.; Schueler, M.; Carninci, P.; Lehrach, H.; Hayashizaki, Y.; Huber, W.; Sperling, S. Genomic organization of transcriptomes in mammals: Coregulation and cofunctionality. Genomics 2007, 89, 580–587. [Google Scholar] [CrossRef] [Green Version]
- Sémon, M.; Duret, L. Evolutionary Origin and Maintenance of Coexpressed Gene Clusters in Mammals. Mol. Biol. Evol. 2006, 23, 1715–1723. [Google Scholar] [CrossRef] [Green Version]
- Le Dily, F.; Baù, D.; Pohl, A.; Vicent, G.P.; Serra, F.; Soronellas, D.; Castellano, G.; Wright, R.H.; Ballare, C.; Filion, G.; et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014, 28, 2151–2162. [Google Scholar] [CrossRef]
- Le Dily, F.; Beato, M. TADs as modular and dynamic units for gene regulation by hormones. FEBS Lett. 2015, 589, 2885–2892. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Chronis, C.; Kronenberg, M.; Chen, X.-F.; Su, T.; Lay, F.D.; Plath, K.; Kurdistani, S.K.; Carey, M.F. Promoter-Enhancer Communication Occurs Primarily within Insulated Neighborhoods. Mol. Cell 2019, 73, 250–263.e5. [Google Scholar] [CrossRef] [Green Version]
- Soler-Oliva, M.E.; Guerrero-Martínez, J.A.; Bachetti, V.; Reyes, J.C. Analysis of the relationship between coexpression domains and chromatin 3D organization. PLoS Comput. Biol. 2017, 13, e1005708. [Google Scholar] [CrossRef] [Green Version]
- Storz, J.F.; Opazo, J.C.; Hoffmann, F.G. Gene duplication, genome duplication, and the functional diversification of vertebrate globins. Mol. Phylogenetics Evol. 2013, 66, 469–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardison, R.C. Globin genes on the move. J. Biol. 2008, 7, 35. [Google Scholar] [CrossRef] [Green Version]
- Hardison, R.C. A brief history of hemoglobins: Plant, animal, protist, and bacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 5675–5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blank, M.; Wollberg, J.; Gerlach, F.; Reimann, K.; Roesner, A.; Hankeln, T.; Fago, A.; Weber, R.E.; Burmester, T. A Membrane-Bound Vertebrate Globin. PLoS ONE 2011, 6, e25292. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970; ISBN 978-3-642-86661-6. [Google Scholar]
- Burmester, T.; Hankeln, T. Function and evolution of vertebrate globins. Acta Physiol. 2014, 211, 501–514. [Google Scholar] [CrossRef]
- Schwarze, K.; Burmester, T. Conservation of globin genes in the “living fossil” Latimeria chalumnae and reconstruction of the evolution of the vertebrate globin family. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 1801–1812. [Google Scholar] [CrossRef]
- Lechauve, C.; Jager, M.; Laguerre, L.; Kiger, L.; Correc, G.; Leroux, C.; Vinogradov, S.; Czjzek, M.; Marden, M.C.; Bailly, X. Neuroglobins, Pivotal Proteins Associated with Emerging Neural Systems and Precursors of Metazoan Globin Diversity. J. Biol. Chem. 2013, 288, 6957–6967. [Google Scholar] [CrossRef] [Green Version]
- Braasch, I.; Brunet, F.; Volff, J.-N.; Schartl, M. Pigmentation Pathway Evolution after Whole-Genome Duplication in Fish. Genome Biol. Evol. 2009, 1, 479–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braasch, I.; Volff, J.-N.; Schartl, M. The Endothelin System: Evolution of Vertebrate-Specific Ligand-Receptor Interactions by Three Rounds of Genome Duplication. Mol. Biol. Evol. 2009, 26, 783–799. [Google Scholar] [CrossRef] [Green Version]
- Holland, P.; Garcia-Fernàndez, J.; Williams, N.A.; Sidow, A. Gene duplications and the origins of vertebrate development. Development. 1994, 1994, 125–133. [Google Scholar] [CrossRef]
- Meyer, A. Hox gene variation and evolution. Nat. Cell Biol. 1998, 391, 227–228. [Google Scholar] [CrossRef] [Green Version]
- Meyer, A.; Schartl, M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999, 11, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Shimeld, S.M.; Holland, P. Vertebrate innovations. Proc. Natl. Acad. Sci. USA 2000, 97, 4449–4452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, H.; Makabe, K. Genome duplications of early vertebrates as a possible chronicle of the evolutionary history of the neural crest. Int. J. Biol. Sci. 2006, 2, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Cohn, M.J. Genome duplication and the origin of the vertebrate skeleton. Curr. Opin. Genet. Dev. 2008, 18, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.G.; Opazo, J.C.; Storz, J.F. Whole-Genome Duplications Spurred the Functional Diversification of the Globin Gene Superfamily in Vertebrates. Mol. Biol. Evol. 2011, 29, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storz, J.F. Gene Duplication and Evolutionary Innovations in Hemoglobin-Oxygen Transport. Physiology 2016, 31, 223–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, M.; Moore, G.W.; Barnabas, J.; Matsuda, G. The phylogeny of human globin genes investigated by the maximum parsimony method. J. Mol. Evol. 1974, 3, 1–48. [Google Scholar] [CrossRef]
- Jeffreys, A.J.; Wilson, V.; Wood, D.; Simons, J.P.; Kay, R.M.; Williams, J.G. Linkage of adult alpha- and beta-globin genes in X. laevis and gene duplication by tetraploidization. Cell 1980, 21, 555–564. [Google Scholar] [CrossRef]
- Hosbach, H.A.; Wyler, T.; Weber, R. The xenopus laevis globin gene family: Chromosomal arrangement and gene structure. Cell 1983, 32, 45–53. [Google Scholar] [CrossRef]
- Patient, R.K.; Elkington, J.A.; Kay, R.M.; Williams, J.G. Internal organization of the major adult α- and β-globin genes of X. laevis. Cell 1980, 21, 565–573. [Google Scholar] [CrossRef]
- Queiroz, J.P.F.; Lima, N.C.B.; Rocha, B.A.M. The rise and fall of globins in the amphibia. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 37, 100759. [Google Scholar] [CrossRef]
- Kovina, A.P.; Petrova, N.; Gushchanskaya, E.S.; Dolgushin, K.V.; Gerasimov, E.; Galitsyna, A.A.; Penin, A.A.; Flyamer, I.; Ioudinkova, E.S.; Gavrilov, A.A.; et al. Evolution of the Genome 3D Organization: Comparison of Fused and Segregated Globin Gene Clusters. Mol. Biol. Evol. 2017, 34, 1492–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, V.S.; Cooper, S.J.; E Deakin, J.; Fulton, B.; Graves, T.; Warren, W.C.; Wilson, R.K.; Graves, J.A. Platypus globin genes and flanking loci suggest a new insertional model for beta-globin evolution in birds and mammals. BMC Biol. 2008, 6, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, V.S.; Ezaz, T.; Deakin, J.E.; Graves, J.A.M. Globin gene structure in a reptile supports the transpositional model for amniote α- and β-globin gene evolution. Chromosom. Res. 2010, 18, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Flint, J.; Tufarelli, C.; Peden, J.; Clark, K.; Daniels, R.J.; Hardison, R.; Miller, W.; Philipsen, S.; Tan-Un, K.C.; McMorrow, T.; et al. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum. Mol. Genet. 2001, 10, 371–382. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, F.G.; Vandewege, M.W.; Storz, J.F.; Opazo, J.C. Gene Turnover and Diversification of the α- and β-Globin Gene Families in Sauropsid Vertebrates. Genome Biol. Evol. 2018, 10, 344–358. [Google Scholar] [CrossRef] [Green Version]
- Opazo, J.C.; Hoffmann, F.G.; Storz, J.F. Genomic evidence for independent origins of -like globin genes in monotremes and therian mammals. Proc. Natl. Acad. Sci. USA 2008, 105, 1590–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, D.; Hope, R.M.; Cooper, S.J.; Gooley, A.A.; Holland, R.A. Linkage of the beta-like omega-globin gene to alpha-like globin genes in an Australian marsupial supports the chromosome duplication model for separation of globin gene clusters. J. Mol. Evol 2004, 58, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Rodin, S.N.; Parkhomchuk, D.V.; Rodin, A.S.; Holmquist, G.P.; Riggs, A.D. Repositioning-Dependent Fate of Duplicate Genes. DNA Cell Biol. 2005, 24, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Philonenko, E.S.; Klochkov, D.B.; Borunova, V.V.; Gavrilov, A.A.; Razin, S.V.; Iarovaia, O.V. TMEM8—A non-globin gene entrapped in the globin web. Nucleic Acids Res. 2009, 37, 7394–7406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prioleau, M.-N.; Nony, P.; Simpson, M.; Felsenfeld, G. An insulator element and condensed chromatin region separate the chicken b-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999, 18, 4035–4048. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razin, S.V.; Ioudinkova, E.S.; Kantidze, O.L.; Iarovaia, O.V. Co-Regulated Genes and Gene Clusters. Genes 2021, 12, 907. https://doi.org/10.3390/genes12060907
Razin SV, Ioudinkova ES, Kantidze OL, Iarovaia OV. Co-Regulated Genes and Gene Clusters. Genes. 2021; 12(6):907. https://doi.org/10.3390/genes12060907
Chicago/Turabian StyleRazin, Sergey V., Elena S. Ioudinkova, Omar L. Kantidze, and Olga V. Iarovaia. 2021. "Co-Regulated Genes and Gene Clusters" Genes 12, no. 6: 907. https://doi.org/10.3390/genes12060907





