Non-Specific Immunity Associated Gut Microbiome in Aristichthys nobilis under Different Rearing Strategies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Designs and Fish Management
2.2. Sample Collection and Pre-Processing
2.3. Non-Specific Immunity Related Enzyme Index Determination
2.4. DNA Extraction, PCR, and Sequencing
2.5. Statistical and Bioinformatics Analysis
3. Result
3.1. Biochemical Parameters Related to Enzyme Activity
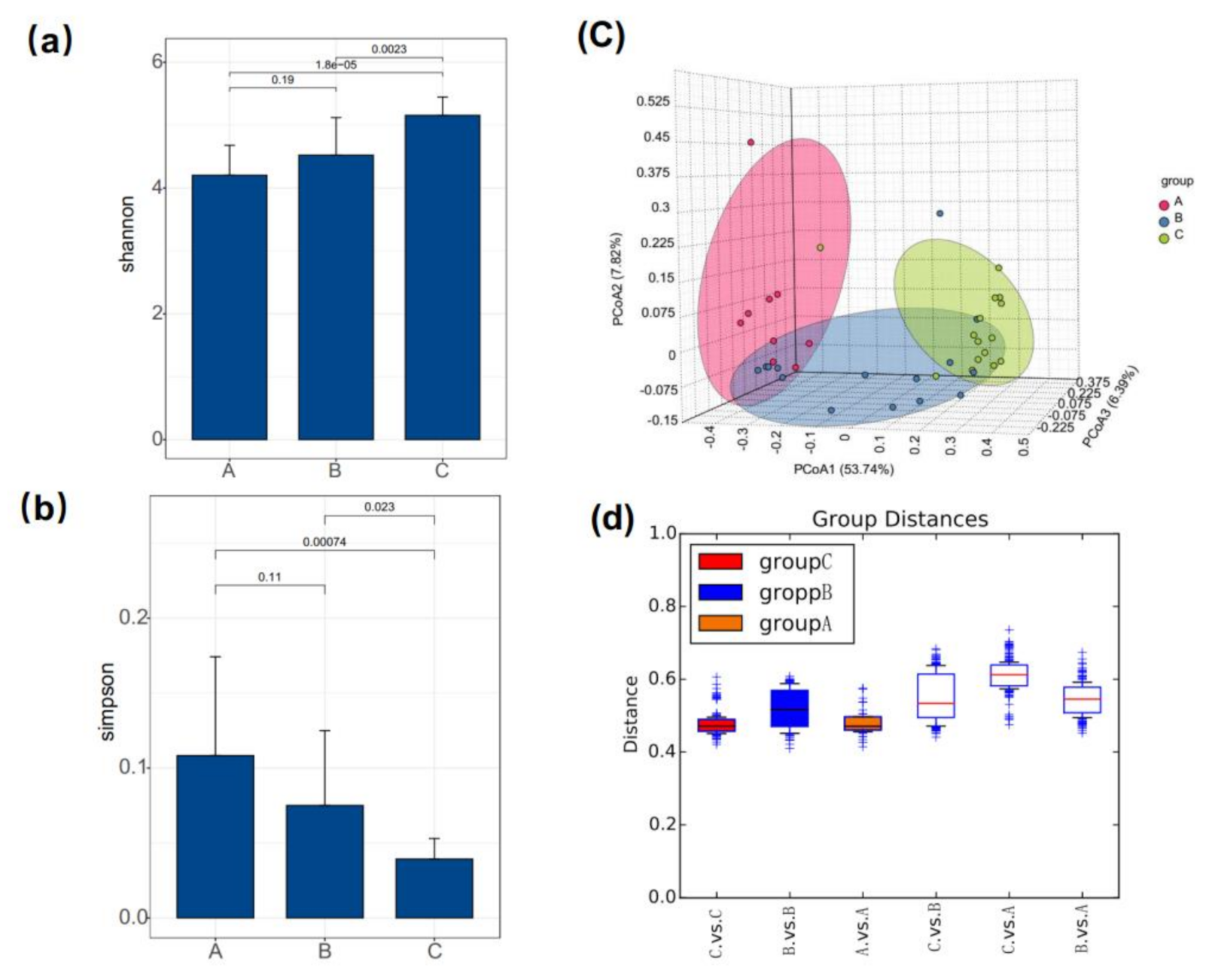
3.2. Gut Bacterial Diversity
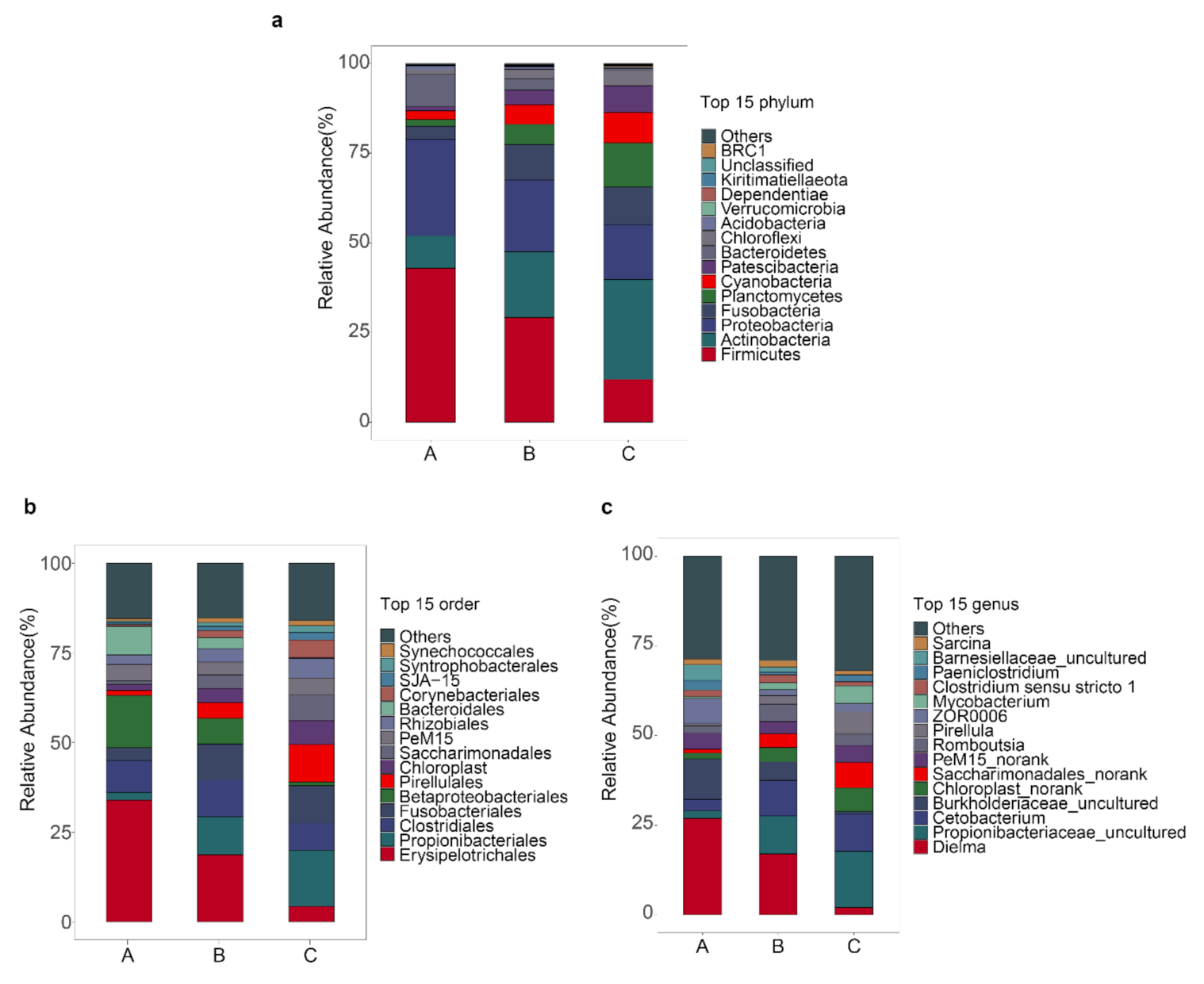
3.3. Gut Bacterial Composition
3.4. Differential Bacterial Composition
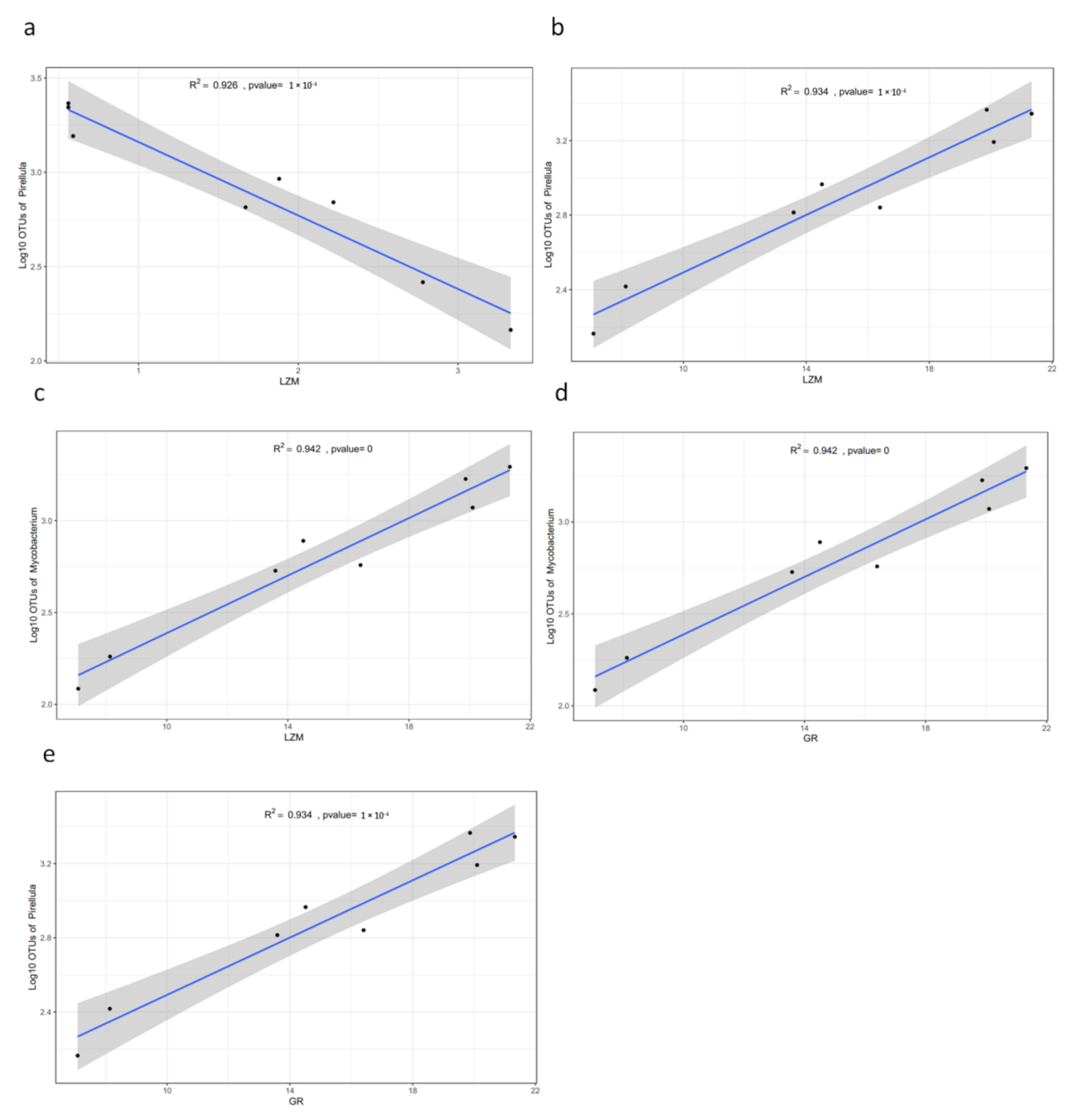
3.5. Functional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, S.; Li, D. Comparative studies on the feeding selectivity of silver carp Hypophthalmichthys molitrix and bighead carp Aristichthys nobilis. J. Fish Biol. 1994, 44, 621–626. [Google Scholar] [CrossRef]
- Anderson, K.R.; Chapman, D.C.; Hayer, C.A. Assessment of dreissenid biodeposits as a potential food resource for invasive Asian carp. Bioinvasions Rec. 2016, 5, 251–257. [Google Scholar] [CrossRef]
- Mi, H.; Wen, Y.; Ge, X.; Sui, R.; Liao, L.; Li, B.; Ren, M.; Zeng, B. The aquaculture situation and development trend of bighead carp in the Pearl River Delta. Sci. Fish Farming 2016, 32, 82–84. [Google Scholar]
- Huang, A. The technology of breeding bighead with compound feed. Fish. Guide Rich 2008, 000, 25–26. [Google Scholar]
- Jorgensen, L.V.G. Zebrafish as a Model for Fish Diseases in Aquaculture. Pathogens 2020, 9, 609. [Google Scholar] [CrossRef]
- Vine, N.G.; Leukes, W.D.; Horst, K. Probiotics in marine larviculture. FEMS Microbiol. Rev. 2006, 30, 404–427. [Google Scholar] [CrossRef] [Green Version]
- Rengpipat, S.; Rukpratanporn, S.; Piyatiratitivorakul, S.; Menasaveta, P. Immunity enhancement in black tiger shrimp (Penaeus monodon) by a probiont bacterium (Bacillus S11). Aquaculture 2000, 191, 271–288. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, J. The mechanism and application of feed lactic acid bacteria probiotics. Chin. J. Anim. Nutr. 2002, 4, 13–18. [Google Scholar]
- Son, V.M.; Chang, C.C.; Wu, M.C.; Guu, Y.K.; Chiu, C.H.; Cheng, W. Dietary administration of the probiotic, Lactobacillus plantarum, enhanced the growth, innate immune responses, and disease resistance of the grouper Epinephelus coioides. Fish Shellfish Immunol. 2009, 26, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.M.; Abdel-Galil Ahmed, Y.; Abdel-Aziz Ghareeb, A.; Mohamed, M.F. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 2008, 25, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Suzer, C.; Çoban, D.; Kamaci, H.O.; Saka, Ş.; Firat, K.; Otgucuoğlu, Ö.; Küçüksari, H. Lactobacillus spp. bacteria as probiotics in gilthead sea bream (Sparus aurata, L.) larvae: Effects on growth performance and digestive enzyme activities. Aquaculture 2008, 280, 140–145. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [Green Version]
- Perez, T.; Balcazar, J.L.; Ruiz-Zarzuela, I.; Halaihel, N.; Vendrell, D.; de Blas, I.; Muzquiz, J.L. Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010, 3, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Radovanović, T.; Borković-Mitić, S.; Perendija, B.R.; Despotović, S.; Saičić, Z. Superoxide dismutase and catalase activities in the liver and muscle of barbel (Barbus barbus) and its intestinal parasite (Pomphoryinchus laevis) from the Danube river, Serbia. Arch. Biol. Sci. 2014, 62, 97–105. [Google Scholar] [CrossRef]
- Paolicchi, F.; Perea, J.; Cseh, S.; Morsella, C. Relationship between Paratuberculosis and the microelements Copper, Zinc, Iron, Selenium and Molybdenum in Beef Cattle. Braz. J. Microbiol. 2013, 44, 153–160. [Google Scholar] [CrossRef]
- Saurabh, S.; Sahoo, P.K. Lysozyme: An important defence molecule of fish innate immune system. Aquac. Res. 2010, 39, 223–239. [Google Scholar] [CrossRef]
- Gajardo, K.; Rodiles, A.; Kortner, T.M.; Krogdahl, A.; Bakke, A.M.; Merrifield, D.L.; Sorum, H. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): A basis for comparative gut microbial research. Sci. Rep. 2016, 6, 30893. [Google Scholar] [CrossRef] [Green Version]
- Gatesoupe, F.J. Live yeasts in the gut: Natural occurrence, dietary introduction, and their effects on fish health and development. Aquaculture 2007, 267, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Roy, R.N.; Sen, S.K.; Ray, A.K. Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquac. Res. 2010, 37, 380–388. [Google Scholar] [CrossRef]
- Torsvik, V.; Ovreas, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Fjellheim, A.J.; Playfoot, K.J.; Skjermo, J.; Vadstein, O. Vibrionaceae dominates the microflora antagonistic towards Listonella anguillarum in the intestine of cultured Atlantic cod (Gadus morhua L.) larvae. Aquaculture 2007, 269, 98–106. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.; Yang, Y.; Xiang, H.; Li, S.; Liang, L.; Sui, H.; Zhan, L.; Lu, X. Deciphering bacterial community changes in zucker diabetic fatty rats based on 16S rRNA gene sequences analysis. Oncotarget 2016, 7, 48941–48952. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Li, A.; Li, M.; Hou, B. Effect of pH on biodiesel production and the microbial structure of glucose-fed activated sludge. Int. Biodeterior. Biodegrad. 2015, 104, 224–230. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011, 77, 3219–3226. [Google Scholar] [CrossRef] [Green Version]
- Westcott, S.L.; Schloss, P.D. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. Peer J. 2015, 3, e1487. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Ijaz, M.U.; Ahmed, M.I.; Zou, X.; Hussain, M.; Zhang, M.; Zhao, F.; Xu, X.; Zhou, G.; Li, C. Beef, Casein, and Soy Proteins Differentially Affect Lipid Metabolism, Triglycerides Accumulation and Gut Microbiota of High-Fat Diet-Fed C57BL/6J Mice. Front. Microbiol. 2018, 9, 2200. [Google Scholar] [CrossRef]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Long, M.; Ji, C.; Shen, Z.; Gatesoupe, F.J.; Zhang, X.; Zhang, Q.; Zhang, L.; Zhao, Y.; Liu, X.; et al. Alterations of the gut microbiome of largemouth bronze gudgeon (Coreius guichenoti) suffering from furunculosis. Sci. Rep. 2016, 6, 30606. [Google Scholar] [CrossRef]
- Stagaman, K.; Burns, A.R.; Guillemin, K.; Bohannan, B.J. The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J. 2017, 11, 1630–1639. [Google Scholar] [CrossRef]
- Balcázar, J.L.; De, B.I.; Ruiz-Zarzuela, I.; Cunningham, D.; Vendrell, D.; Múzquiz, J.L. The role of probiotics in aquaculture. Vet. Microbiol. 2006, 114, 173–186. [Google Scholar] [CrossRef]
- Lin, X.; Xia, Y.; Wang, G.; Yang, Y.; Xiong, Z.; Lv, F.; Zhou, W.; Ai, L. Lactic Acid Bacteria With Antioxidant Activities Alleviating Oxidized Oil Induced Hepatic Injury in Mice. Front. Microbiol. 2018, 9, 2684. [Google Scholar] [CrossRef] [PubMed]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; Gonzalez, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Chen, C.; Jia, L.; He, X.; Zhang, B. Comparison of the intestinal microbiota composition and function in healthy and diseased Yunlong Grouper. AMB Express 2019, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef] [Green Version]
- Estruch, G.; Collado, M.C.; Penaranda, D.S.; Tomas Vidal, A.; Jover Cerda, M.; Perez Martinez, G.; Martinez-Llorens, S. Impact of Fishmeal Replacement in Diets for Gilthead Sea Bream (Sparus aurata) on the Gastrointestinal Microbiota Determined by Pyrosequencing the 16S rRNA Gene. PLoS ONE 2015, 10, e0136389. [Google Scholar] [CrossRef]
- Ricaud, K.; Rey, M.; Plagnes-Juan, E.; Larroquet, L.; Even, M.; Quillet, E.; Skiba-Cassy, S.; Panserat, S. Composition of Intestinal Microbiota in Two Lines of Rainbow Trout (Oncorhynchus Mykiss) Divergently Selected for Muscle Fat Content. Open Microbiol. J. 2018, 12, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Penn, K.; Jenkins, C.; Nett, M.; Udwary, D.W.; Gontang, E.A.; McGlinchey, R.P.; Foster, B.; Lapidus, A.; Podell, S.; Allen, E.E.; et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009, 3, 1193–1203. [Google Scholar] [CrossRef]
- Mendes, M.C.S.; Paulino, D.S.; Brambilla, S.R.; Camargo, J.A.; Persinoti, G.F.; Carvalheira, J.B.C. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J. Gastroenterol. 2018, 24, 1995–2008. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Hamilton, M.J.; Staley, C.; Sadowsky, M.J.; Sorensen, P.W. Environment shapes the fecal microbiome of invasive carp species. Microbiome 2016, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- Finegold, S.M.; Vaisanen, M.L.; Molitoris, D.R.; Tomzynski, T.J.; Song, Y.; Liu, C.; Collins, M.D.; Lawson, P.A. Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst. Appl. Microbiol. 2003, 26, 177–181. [Google Scholar] [CrossRef]
- Sugita, H.; Miyajima, C.; Deguchi, Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture 1991, 92, 267–276. [Google Scholar] [CrossRef]

| Characteristics/Tissue | Index | Group A (n = 9) | Group B (n = 15) | Group C (n = 15) | p-Value |
|---|---|---|---|---|---|
| Characteristics | Initial weight (g) | 47.57 ± 6.08 | 52.40 ± 1.55 | 51.87 ± 2.13 | NS |
| Final weight (g) | 77.85 ± 11.86 | 121.80 ± 2.90 | 120.54 ± 5.83 | <0.01 | |
| Weight gain rate (%) | 63.40 ± 4.04 | 132.46 ± 1.75 | 132.34 ± 1.67 | <0.001 | |
| Growth rate (%) | 50.47 ± 9.63 | 115.66 ± 2.37 | 114.44 ± 6.16 | <0.001 | |
| Survival rate (%) | 100 | 100 | 100 | NS | |
| Fatness (%) | 1.97 ± 0.09 | 2.00 ± 0.09 | 2.00 ± 0.08 | NS | |
| Visceral body ratio (%) | 3.77 ± 0.54 | 6.17 ± 0.74 | 6.14 ± 0.64 | <0.01 | |
| Hepatopancreas | LZM (ug/mL) | 3.06 ± 0.39 a | 1.92 ± 0.28 b | 0.57 ± 0.02 c | <0.001 |
| CAT (U/mgprot) | 13.75 ± 2.37 | 11.00 ± 0.65 | 11.86 ± 0.34 | NS | |
| GR (U/gprot) | 18.36 ± 3.84 | 15.35 ± 3.72 | 18.46 ± 1.01 | NS | |
| GSH-PX (U/mgprot) | 1182.86 ± 65.99 a | 1162.77 ± 35.16 a | 760.89 ± 49.84 b | <0.001 | |
| SOD (U/mgprot) | 34.85 ± 2.58 c | 130.29 ± 8.76 a | 86.57 ± 2.98 b | <0.001 | |
| Spleen | LZM (ug/mL) | 14.66 ± 0.31 b | 16.82 ± 1.14 b | 22.27 ± 2.22 a | <0.01 |
| CAT (U/mgprot) | 1.11 ± 0.06 b | 2.84 ± 0.30 a | 1.07 ± 0.29 b | <0.001 | |
| GR (U/gprot) | 7.61 ± 0.75 c | 14.83 ± 1.43 b | 20.44 ± 0.78 a | <0.001 | |
| GSH-PX (U/mgprot) | 670.89 ± 27.66 a | 449.05 ± 38.08 b | 452.16 ± 15.25 b | <0.001 | |
| SOD (U/mgprot) | 381.52 ± 0.99 b | 449.27 ± 14.30 a | 450.89 ± 18.71 a | <0.01 |
| Group | OTUs | Chao | Shannon | Simpson | Coverage |
|---|---|---|---|---|---|
| A | 1806 +/− 267 | 3111.11 +/− 543.80 | 4.20 +/− 0.47 | 0.1084 +/− 0.0659 | 0.9862 +/− 0.0029 |
| B | 1764 +/− 303 | 2844.00 +/− 562.98 | 4.52+/− 0.60 | 0.075 +/− 0.0499 | 0.9871 +/− 0.0027 |
| C | 2298 +/− 196 | 3739.67 +/− 315.29 | 5.16 +/− 0.291 | 0.0393 +/− 0.0137 | 0.9837 +/− 0.0026 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, J.; Wang, Z.; Wang, B.; Mei, H.; Zhai, X.; Zhuang, Z.; Chen, M.; Zhang, Y. Non-Specific Immunity Associated Gut Microbiome in Aristichthys nobilis under Different Rearing Strategies. Genes 2021, 12, 916. https://doi.org/10.3390/genes12060916
Yuan J, Wang Z, Wang B, Mei H, Zhai X, Zhuang Z, Chen M, Zhang Y. Non-Specific Immunity Associated Gut Microbiome in Aristichthys nobilis under Different Rearing Strategies. Genes. 2021; 12(6):916. https://doi.org/10.3390/genes12060916
Chicago/Turabian StyleYuan, Jianming, Zhijian Wang, Bo Wang, Huiqing Mei, Xuliang Zhai, Zhenhua Zhuang, Maoshan Chen, and Yaoguang Zhang. 2021. "Non-Specific Immunity Associated Gut Microbiome in Aristichthys nobilis under Different Rearing Strategies" Genes 12, no. 6: 916. https://doi.org/10.3390/genes12060916
APA StyleYuan, J., Wang, Z., Wang, B., Mei, H., Zhai, X., Zhuang, Z., Chen, M., & Zhang, Y. (2021). Non-Specific Immunity Associated Gut Microbiome in Aristichthys nobilis under Different Rearing Strategies. Genes, 12(6), 916. https://doi.org/10.3390/genes12060916






