GWAS Hits for Bilateral Convergent Strabismus with Exophthalmos in Holstein Cattle Using Imputed Sequence Level Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Clinical Phenotypes
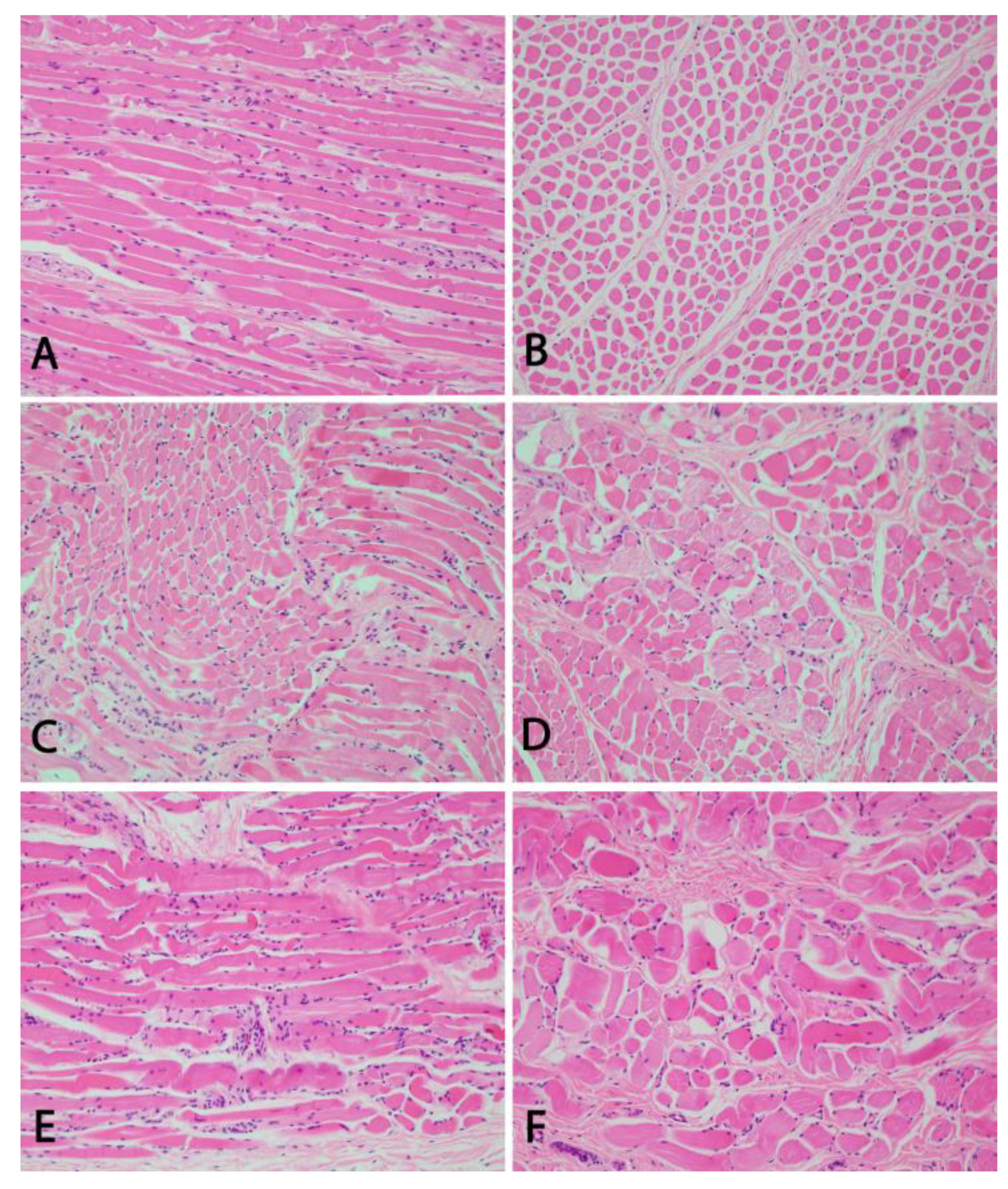
2.2. Histo- and Neuropathological Examinations
2.3. SNP Genotyping, Imputation and Genome-Wide Association Study (GWAS)
2.4. Evaluation of the Molecular Consequences of Amino Acid Substitutions
3. Results
3.1. Clinical Phenotype
3.2. Pathological Phenotype
3.3. Genome-Wide Association Study and Candiate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Distl, O.; Wenninger, A.; Kräusslich, H. Inheritance of convergent strabismus with exophthalmus in cattle. Dtsch. Tierärztliche Wochenschr. 1991, 98, 354–356. [Google Scholar]
- Regan, W.M.; Gregory, P.W.; Mead, S.W. Hereditary strabismus in Jersey cattle. J. Hered. 1944, 35, 233–234. [Google Scholar] [CrossRef]
- Schütz-Hänke, W.; Stöber, M.; Drommer, W. Klinische, genealogische und pathomorphologische Untersuchungen an schwarzbunten Rindern mit beiderseitigem exophthalmisch-konvergierendem Schielen. Dtsch. Tierärztliche Wochenschr. 1979, 86, 185–191. [Google Scholar]
- Gerst, M.; Distl, O. Einflüsse auf die Dissemination des bilateralen Strabismus convergens mit Exophthalmus beim Rind. Arch. Für Tierz. 1997, 40, 401–412. [Google Scholar]
- Mömke, S.; Fink, S.; Wöhlke, A.; Drögemüller, C.; Distl, O. Linkage of bilateral convergent strabismus with exophthalmus (BCSE) to BTA5 and BTA18 in German Brown cattle. Anim. Genet. 2008, 39, 544–549. [Google Scholar] [CrossRef]
- Distl, O.; Gerst, M. Association Analysis between Bilateral Convergent Strabismus with Exophthalmus and Milk Production Traits in Dairy Cattle. J. Vet. Med. Ser. A 2000, 47, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Vogt, C. Untersuchungen zum bilateralen Strabismus convergens mit Exophthalmus (BCSE) beim Deutschen Braunvieh. Ph.D. Thesis, University of Veterinary Medicine Hannover, Hannover, Germany, 2000. [Google Scholar]
- Distl, O. Analysis of pedigrees in dairy cattle segregating for bilateral strabismus with exophthalmus. J. Anim. Breed. Genet. 1993, 110, 393–400. [Google Scholar] [CrossRef]
- Mömke, S.; Distl, O. Bilateral convergent strabismus with exophthalmus (BCSE) in cattle: An overview of clinical signs and genetic traits. Vet. J. 2007, 173, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.; Mömke, S.; Distl, O. PLXNC1 and RDH13 associated with bilateral convergent strabismus with exophthalmus in German Brown cattle. Mol. Vis. 2012, 18, 2229–2240. [Google Scholar] [PubMed]
- McClelland, C.; Manousakis, G.; Lee, M.S. Progressive External Ophthalmoplegia. Curr. Neurol. Neurosci. Rep. 2016, 16, 53. [Google Scholar] [CrossRef]
- Hauke, G. Candidate Gene Analysis for Bilateral Convergent Strabismus with Exophthalmus in German Brown Cattle. Ph.D. Thesis, University of Veterinary Medicine Hannover, Hannover, Germany, 2003. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Phanstiel, D.H. Sushi: Tools for Visualizing Genomics Data, R package version 1.26.0; 2021. [Google Scholar]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Schoser, B.G.H. Ocular myositis: Diagnostic assessment, differential diagnoses, and therapy of a rare muscle disease—five new cases and review. Clin. Ophthalmol. 2007, 1, 37–42. [Google Scholar]
- Visuttijai, K.; Hedberg-Oldfors, C.; Lindgren, U.; Nordström, S.; Elíasdóttir, Ó.; Lindberg, C.; Oldfors, A. Progressive external ophthalmoplegia associated with novel MT-TN mutations. Acta Neurol. Scand. 2021, 143, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Gelatt, K.N. (Ed.) Essentials of Veterinary Ophthalmology, 3rd ed.; John Wiley & Sons Inc.: Ames, IA, USA, 2014; ISBN 978-1-118-77192-1. [Google Scholar]
- Qanbari, S.; Pimentel, E.C.G.; Tetens, J.; Thaller, G.; Lichtner, P.; Sharifi, A.R.; Simianer, H. The pattern of linkage disequilibrium in German Holstein cattle. Anim. Genet. 2010, 41, 346–356. [Google Scholar] [CrossRef]
- Flury, C.; Tapio, M.; Sonstegard, T.; Drögemüller, C.; Leeb, T.; Simianer, H.; Hanotte, O.; Rieder, S. Effective population size of an indigenous Swiss cattle breed estimated from linkage disequilibrium. J. Anim. Breed. Genet. 2010, 127, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, R.A.; Taylor, J.F.; van Tassell, C.P.; Barendse, W.; Eversole, K.A.; Gill, C.A.; Green, R.D.; Hamernik, D.L.; Kappes, S.M.; Lien, S.; et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 2009, 324, 528–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Binsbergen, R.; Bink, M.C.; Calus, M.P.; van Eeuwijk, F.A.; Hayes, B.J.; Hulsegge, I.; Veerkamp, R.F. Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle. Genet. Sel. Evol. 2014, 46, 41. [Google Scholar] [CrossRef] [Green Version]
- Russel, F.G.; Koenderink, J.B.; Masereeuw, R. Multidrug resistance protein 4 (MRP4/ABCC4): A versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol. Sci. 2008, 29. [Google Scholar] [CrossRef] [PubMed]
- Berthier, J.; Arnion, H.; Saint-Marcoux, F.; Picard, N. Multidrug resistance-associated protein 4 in pharmacology: Overview of its contribution to pharmacokinetics, pharmacodynamics and pharmacogenetics. Life Sci. 2019, 231, 116540. [Google Scholar] [CrossRef] [PubMed]
- Grewal, G.K.; Kukal, S.; Kanojia, N.; Saso, L.; Kukreti, S.; Kukreti, R. Effect of Oxidative Stress on ABC Transporters: Contribution to Epilepsy Pharmacoresistance. Molecules 2017, 22, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumiya, W.; Kusuhara, S.; Hayashibe, K.; Maruyama, K.; Kusuhara, H.; Tagami, M.; Schuetz, J.D.; Negi, A. Forskolin Modifies Retinal Vascular Development in Mrp4-Knockout Mice. Investig. Ophthalmol. Vis. Sci. 2012, 53, 8029–8035. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Kusuhara, S.; Katsuyama-Yoshikawa, A.; Nobuyoshi, S.; Kitamura, M.; Mori, S.; Sotani, N.; Ueda, K.; Matsumiya, W.; Miki, A.; et al. Changes in Gene Expression Profiling and Phenotype in Aged Multidrug Resistance Protein 4-Deficient Mouse Retinas. Antioxidants 2021, 10, 455. [Google Scholar] [CrossRef]
- Mottis, A.; Mouchiroud, L.; Auwerx, J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013, 27, 819–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.D.; Evans, R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 1995, 377, 454–457. [Google Scholar] [CrossRef]
- Jepsen, K.; Solum, D.; Zhou, T.; McEvilly, R.J.; Kim, H.-J.; Glass, C.K.; Hermanson, O.; Rosenfeld, M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 2007, 450, 415–419. [Google Scholar] [CrossRef]
- Park, D.H.; Hong, S.J.; Salinas, R.D.; Liu, S.J.; Sun, S.W.; Sgualdino, J.; Testa, G.; Matzuk, M.M.; Iwamori, N.; Lim, D.A. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014, 8, 1290–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.-C.; Kao, H.-Y.; Mitzutani, A.; Banayo, E.; Rajan, H.; McKeown, M.; Evans, R.M. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 4047–4052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoghbi, H.Y.; Orr, H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000, 23, 217–247. [Google Scholar] [CrossRef] [PubMed]
- Synofzik, M.; Haack, T.B.; Kopajtich, R.; Gorza, M.; Rapaport, D.; Greiner, M.; Schönfeld, C.; Freiberg, C.; Schorr, S.; Holl, R.W.; et al. Absence of BiP co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am. J. Hum. Genet. 2014, 95, 689–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozon, Z.A.; Alikasifoglu, A.; Kandemir, N.; Aydin, B.; Gonc, E.N.; Karaosmanoglu, B.; Celik, N.B.; Eroglu-Ertugrul, N.G.; Taskiran, E.Z.; Haliloglu, G.; et al. Novel insights into diabetes mellitus due to DNAJC3-defect: Evolution of neurological and endocrine phenotype in the pediatric age group. Pediatr. Diabetes 2020, 21, 1176–1182. [Google Scholar] [CrossRef] [PubMed]
- Lytrivi, M.; Senée, V.; Salpea, P.; Fantuzzi, F.; Philippi, A.; Abdulkarim, B.; Sawatani, T.; Marín-Cañas, S.; Pachera, N.; Degavre, A.; et al. DNAJC3 deficiency induces β-cell mitochondrial apoptosis and causes syndromic young-onset diabetes. Eur. J. Endocrinol. 2021, 184, 455–468. [Google Scholar] [CrossRef]

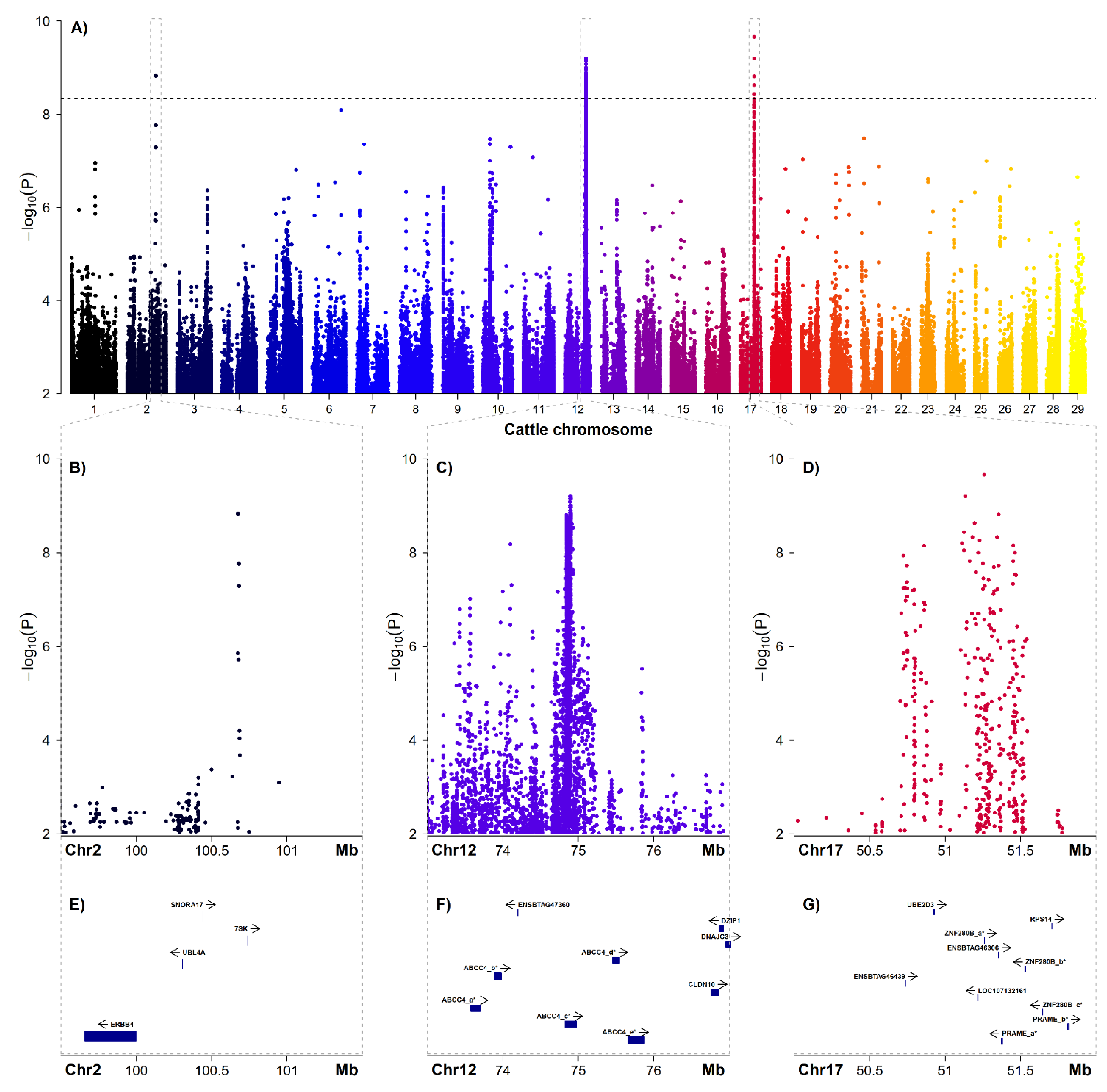
| Chr | rsID | Position [bp] 1 | Alleles 2 | MAF 3 | Odds Ratio | p-Value 4 | VEP 5 | Gene |
|---|---|---|---|---|---|---|---|---|
| 2 | rs136316260 | 100,672,388 | G/T | 0.17 | 38.75 | 1.49E-09 | intergenic | |
| 2 | rs134704382 | 100,674,312 | G/A | 0.17 | 38.75 | 1.49E-09 | intergenic | |
| 2 | rs135186290 | 100,675,331 | A/G | 0.17 | 38.75 | 1.49E-09 | intergenic | |
| 2 | rs134623922 | 100,675,365 | G/A | 0.17 | 38.75 | 1.49E-09 | intergenic | |
| 2 | rs385880764 | 100,675,837 | -/ATC | 0.17 | 38.75 | 1.49E-09 | intergenic | |
| 2 | rs133964128 | 100,676,979 | A/G | 0.17 | 38.75 | 1.49E-09 | intergenic | |
| 12 | rs377992474 | 74,888,457 | T/A | 0.34 | 9.90 | 8.36E-10 | intron | ABCC4 |
| 12 | rs468443940 | 74,891,825 | T/G | 0.36 | 8.76 | 1.28E-09 | intron | ABCC4 |
| 12 | rs381451112 | 74,892,681 | C/G | 0.34 | 10.78 | 1.41E-09 | intron | ABCC4 |
| 12 | rs383448105 | 74,892,723 | A/G | 0.34 | 10.04 | 6.27E-10 | intron | ABCC4 |
| 12 | rs378170580 | 74,892,729 | T/G | 0.34 | 10.84 | 1.00E-09 | intron | ABCC4 |
| 12 | rs384157620 | 74,892,843 | A/G | 0.34 | 9.68 | 1.09E-09 | intron | ABCC4 |
| 12 | rs110869430 | 74,893,319 | T/G | 0.33 | 9.70 | 7.05E-10 | intron | ABCC4 |
| 12 | rs109771712 | 74,893,351 | C/G | 0.32 | 9.64 | 9.94E-10 | intron | ABCC4 |
| 12 | rs876085957 | 74,893,362 | C/T | 0.34 | 9.86 | 1.09E-09 | intron | ABCC4 |
| 12 | rs110191959 | 74,894,971 | A/G | 0.31 | 12.26 | 1.46E-09 | intron | ABCC4 |
| 12 | rs384153179 | 74,895,060 | T/C | 0.32 | 11.94 | 1.55E-09 | intron | ABCC4 |
| 17 | rs451202354 | 51,135,057 | C/T | 0.17 | 153.30 | 6.29E-10 | intergenic | |
| 17 | rs384954012 | 51,259,815 | A/- | 0.17 | 90.24 | 2.17E-10 | intergenic | |
| 17 | rs377905476 | 51,356,292 | G/T | 0.18 | 45.64 | 1.53E-09 | splice region | ENSBTAG00000046306 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bögeholz, A.; Falker-Gieske, C.; Guélat, M.; Gurtner, C.; Hunziker, S.; Oevermann, A.; Thaller, G.; Drögemüller, C.; Tetens, J. GWAS Hits for Bilateral Convergent Strabismus with Exophthalmos in Holstein Cattle Using Imputed Sequence Level Genotypes. Genes 2021, 12, 1039. https://doi.org/10.3390/genes12071039
Bögeholz A, Falker-Gieske C, Guélat M, Gurtner C, Hunziker S, Oevermann A, Thaller G, Drögemüller C, Tetens J. GWAS Hits for Bilateral Convergent Strabismus with Exophthalmos in Holstein Cattle Using Imputed Sequence Level Genotypes. Genes. 2021; 12(7):1039. https://doi.org/10.3390/genes12071039
Chicago/Turabian StyleBögeholz, Anke, Clemens Falker-Gieske, Monika Guélat, Corinne Gurtner, Sibylle Hunziker, Anna Oevermann, Georg Thaller, Cord Drögemüller, and Jens Tetens. 2021. "GWAS Hits for Bilateral Convergent Strabismus with Exophthalmos in Holstein Cattle Using Imputed Sequence Level Genotypes" Genes 12, no. 7: 1039. https://doi.org/10.3390/genes12071039
APA StyleBögeholz, A., Falker-Gieske, C., Guélat, M., Gurtner, C., Hunziker, S., Oevermann, A., Thaller, G., Drögemüller, C., & Tetens, J. (2021). GWAS Hits for Bilateral Convergent Strabismus with Exophthalmos in Holstein Cattle Using Imputed Sequence Level Genotypes. Genes, 12(7), 1039. https://doi.org/10.3390/genes12071039






