Limited Association between Schizophrenia Genetic Risk Factors and Transcriptomic Features
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. S-LDSC Annotation File Generation
2.3. S-LDSC
2.4. TF–Target Gene Linkage Analysis
3. Results
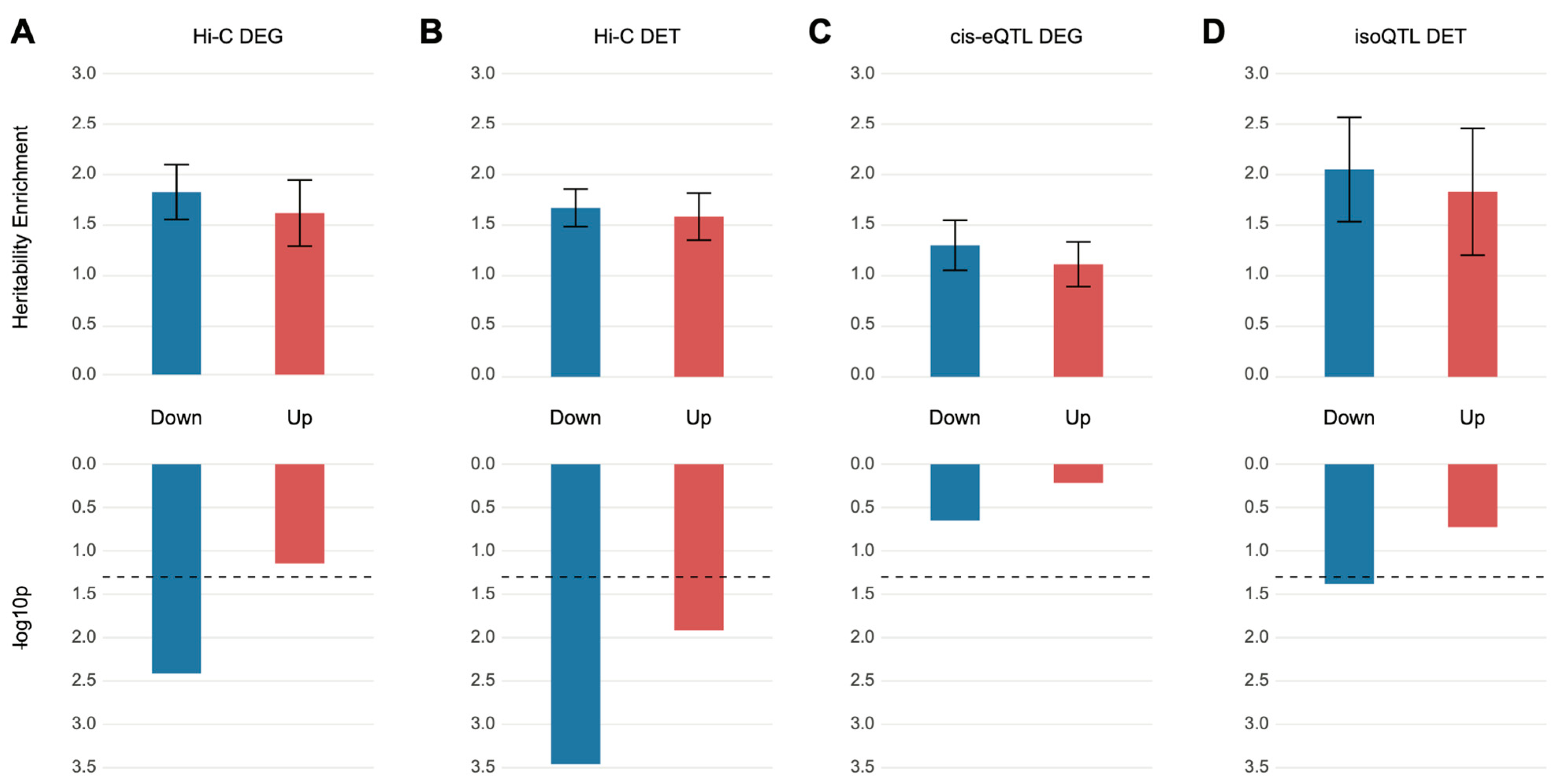
3.1. Differentially Expressed Genes and Transcripts
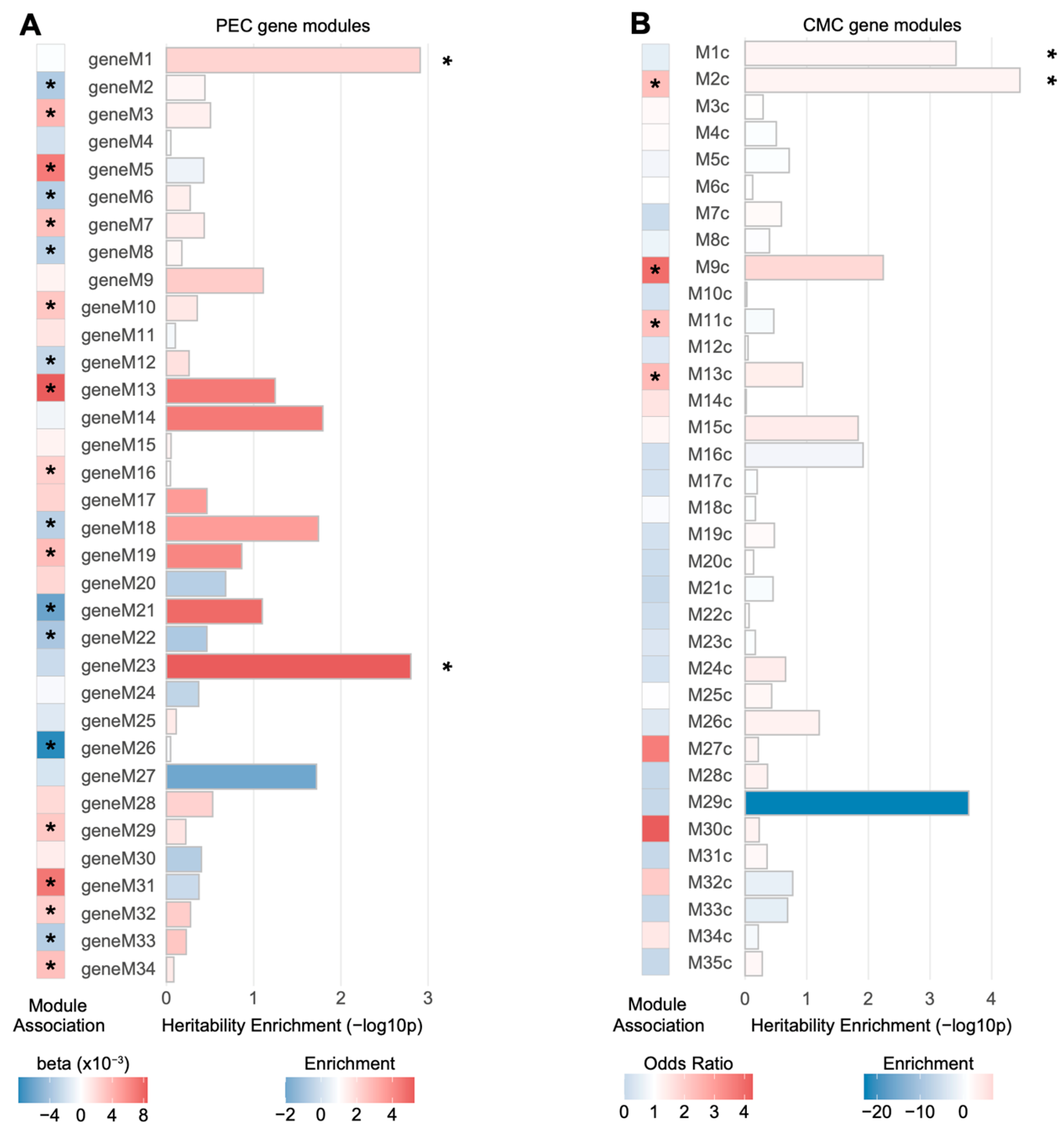
3.2. Co-Expression Networks
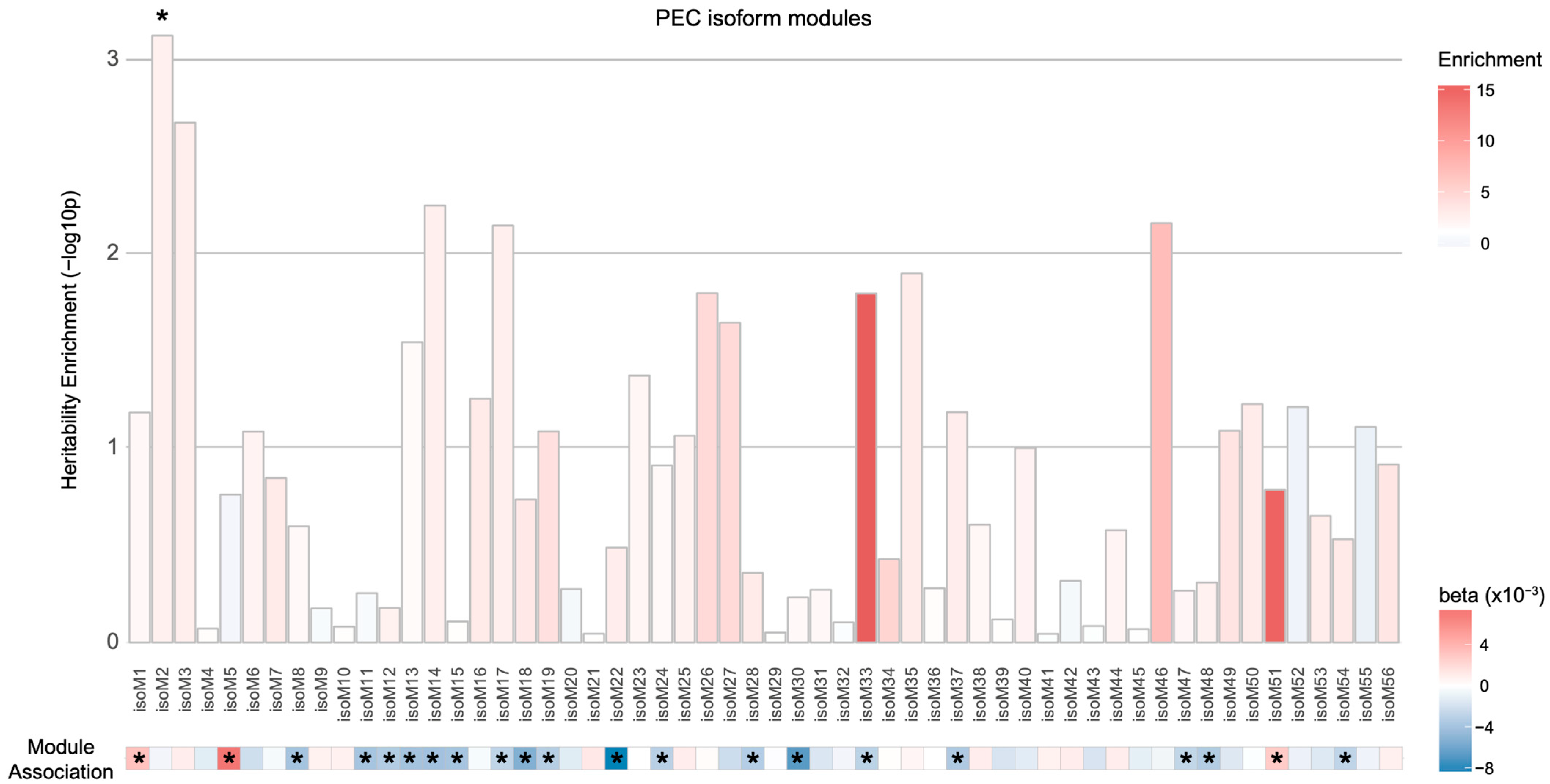
3.3. Isoform-Level Co-Expression Modules
3.4. Network Centrality
3.5. Cell Type-Specific Transcriptomic Signature
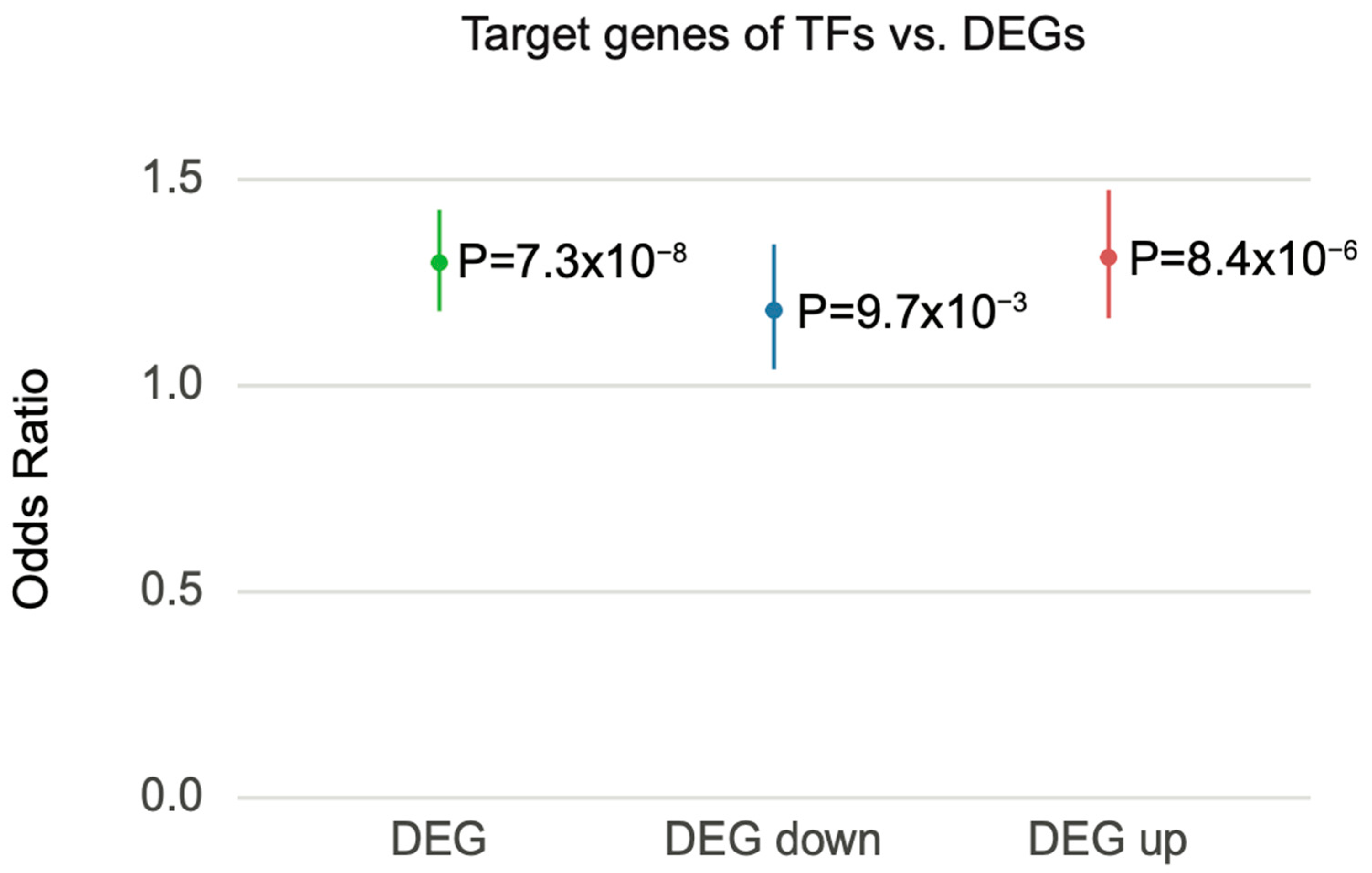
3.6. Genetic Risk Factors Drive Transcriptomic Alterations through the Regulation of Transcription Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pardiñas, A.F.; Holmans, P.; Pocklington, A.J.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.L.; et al. Common Schizophrenia Alleles Are Enriched in Mutation-Intolerant Genes and in Regions under Strong Background Selection. Nat. Genet. 2018, 50, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Ripke, S.; Walters, J.T.R.; O’Donovan, M.C. Mapping Genomic Loci Prioritises Genes and Implicates Synaptic Biology in Schizophrenia. MedRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Qi, G.; Park, J.-H.; Chatterjee, N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet. 2018, 50, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [Green Version]
- Fullard, J.F.; Giambartolomei, C.; Hauberg, M.E.; Xu, K.; Voloudakis, G.; Shao, Z.; Bare, C.; Dudley, J.T.; Mattheisen, M.; Robakis, N.K.; et al. Open Chromatin Profiling of Human Postmortem Brain Infers Functional Roles for Non-Coding Schizophrenia Loci. Hum. Mol. Genet. 2017, 26, 1942–1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, H.; De La Torre-Ubieta, L.; Stein, J.; Parikshak, N.; Huang, J.; Opland, C.K.; Gandal, M.; Sutton, G.J.; Hormozdiari, F.; Lu, D.; et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 2016, 538, 523–527. [Google Scholar] [CrossRef] [Green Version]
- Singh, T.; Kurki, M.I.; Curtis, D.; Purcell, S.M.; Crooks, L.; McRae, J.; Suvisaari, J.; Chheda, H.; Blackwood, D.; Breen, G.; et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat. Neurosci. 2016, 19, 571–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sey, N.Y.A.; Hu, B.; Mah, W.; Fauni, H.; McAfee, J.C.; Rajarajan, P.; Brennand, K.J.; Akbarian, S.; Won, H. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 2020, 23, 583–593. [Google Scholar] [CrossRef]
- Mah, W.; Won, H. The Three-Dimensional Landscape of the Genome in Human Brain Tissue Unveils Regulatory Mechanisms Leading to Schizophrenia Risk. Schizophr. Res. 2020, 217, 17–25. [Google Scholar] [CrossRef]
- Consortium, Network Pathway Analysis Subgroup of the Psychiatric Genomics. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015, 18, 199–209. [Google Scholar] [CrossRef]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; Van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127. [Google Scholar] [CrossRef] [Green Version]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Li, Z.; Warrell, J.; Won, H.; Shi, X.; Navarro, F.C.P.; Clarke, D.; Gu, M.; Emani, P.; Yang, Y.T.; et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 2018, 362, eaat8464. [Google Scholar] [CrossRef] [Green Version]
- Ruzicka, W.B.; Mohammadi, S.; Davila-Velderrain, J.; Subburaju, S.; Tso, D.R.; Hourihan, M.; Kellis, M. Single-cell dissection of schizophrenia reveals neurodevelopmental-synaptic axis and transcriptional resilience. MedRxiv 2020. [Google Scholar] [CrossRef]
- Finucane, H.K.; Bulik-Sullivan, B.; Gusev, A.; Trynka, G.; Reshef, Y.; Loh, P.R.; Anttila, V.; Xu, H.; Zang, C.; Farh, K.; et al. Partitioning Heritability by Functional Annotation Using Genome-Wide Association Summary Statistics. Nat. Genet. 2015, 47, 1228–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pergola, G.; Di Carlo, P.; Jaffe, A.E.; Papalino, M.; Chen, Q.; Hyde, T.M.; Kleinman, J.E.; Shin, J.H.; Rampino, A.; Blasi, G.; et al. Prefrontal Coexpression of Schizophrenia Risk Genes Is Associated with Treatment Response in Patients. Biol. Psychiatry 2019, 86, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Torretta, S.; Rampino, A.; Basso, M.; Pergola, G.; Di Carlo, P.; Shin, J.H.; Kleinman, J.E.; Hyde, T.M.; Weinberger, D.R.; Masellis, R.; et al. NURR1 and ERR1 Modulate the Expression of Genes of a DRD2 Coexpression Network Enriched for Schizophrenia Risk. J. Neurosci. 2020, 40, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Marín, O. Interneuron Dysfunction in Psychiatric Disorders. Nat. Rev. Neurosci. 2012, 13, 107–120. [Google Scholar] [CrossRef]
- Vuong, C.K.; Black, D.L.; Zheng, S. The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 2016, 17, 265–281. [Google Scholar] [CrossRef]
- Kim, S.S.; Dai, C.; Hormozdiari, F.; Van De Geijn, B.; Gazal, S.; Park, Y.; O’Connor, L.; Amariuta, T.; Loh, P.-R.; Finucane, H.; et al. Genes with High Network Connectivity Are Enriched for Disease Heritability. Am. J. Hum. Genet. 2019, 104, 896–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blake, D.J.; Forrest, M.; Chapman, R.M.; Tinsley, C.L.; O’Donovan, M.; Owen, M.J. TCF4, Schizophrenia, and Pitt-Hopkins Syndrome. Schizophr. Bull. 2010, 36, 443–447. [Google Scholar] [CrossRef] [Green Version]
- Xia, H.; Jahr, F.M.; Kim, N.-K.; Xie, L.; Shabalin, A.A.; Bryois, J.; Sweet, D.H.; Kronfol, M.M.; Palasuberniam, P.; McRae, M.; et al. Building a Schizophrenia Genetic Network: Transcription Factor 4 Regulates Genes Involved in Neuronal Development and Schizophrenia Risk. Hum. Mol. Genet. 2018, 27, 3246–3256. [Google Scholar] [CrossRef]
- Torshizi, A.D.; Armoskus, C.; Zhang, H.; Forrest, M.P.; Zhang, S.; Souaiaia, T.; Evgrafov, O.V.; Knowles, J.A.; Duan, J.; Wang, K. Deconvolution of transcriptional networks identifies TCF4 as a master regulator in schizophrenia. Sci. Adv. 2019, 5, eaau4139. [Google Scholar] [CrossRef] [Green Version]
- Boldog, E.; Bakken, T.E.; Hodge, R.D.; Novotny, M.; Aevermann, B.D.; Baka, J.; Bordé, S.; Close, J.L.; Diez-Fuertes, F.; Ding, S.-L.; et al. Transcriptomic and Morphophysiological Evidence for a Specialized Human Cortical GABAergic Cell Type. Nat. Neurosci. 2018, 21, 1185–1195. [Google Scholar] [CrossRef]
- Spiess, K.; Won, H. Regulatory landscape in brain development and disease. Curr. Opin. Genet. Dev. 2020, 65, 53–60. [Google Scholar] [CrossRef]
- Won, H.; Huang, J.; Opland, C.K.; Hartl, C.L.; Geschwind, D.H. Human Evolved Regulatory Elements Modulate Genes Involved in Cortical Expansion and Neurodevelopmental Disease Susceptibility. Nat. Commun. 2019, 10, 2396. [Google Scholar] [CrossRef] [Green Version]
- Enwright, J.F., III; Huo, Z.; Arion, D.; Corradi, J.P.; Tseng, G.; Lewis, D.A. Transcriptome Alterations of Prefrontal Cortical Parvalbumin Neurons in Schizophrenia. Mol. Psychiatry 2018, 23, 1606–1613. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.A.; Curley, A.A.; Glausier, J.R.; Volk, D.W. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012, 35, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Won, H.; Mah, W.; Park, R.; Kassim, B.; Spiess, K.; Kozlenkov, A.; Crowley, C.A.; Pochareddy, S.; Li, Y.; et al. Neuronal and glial 3D chromatin architecture informs the cellular etiology of brain disorders. Nat. Commun. 2021, 12, 3968. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.I.; Pritchard, J.K. Trans Effects on Gene Expression Can Drive Omnigenic Inheritance. Cell 2019, 177, 1022–1034.e6. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, A.W.; Peery, J.D.; Won, H. Limited Association between Schizophrenia Genetic Risk Factors and Transcriptomic Features. Genes 2021, 12, 1062. https://doi.org/10.3390/genes12071062
Yu AW, Peery JD, Won H. Limited Association between Schizophrenia Genetic Risk Factors and Transcriptomic Features. Genes. 2021; 12(7):1062. https://doi.org/10.3390/genes12071062
Chicago/Turabian StyleYu, Alice W., J. David Peery, and Hyejung Won. 2021. "Limited Association between Schizophrenia Genetic Risk Factors and Transcriptomic Features" Genes 12, no. 7: 1062. https://doi.org/10.3390/genes12071062








