How Important Are Structural Variants for Speciation?
Abstract
1. Introduction
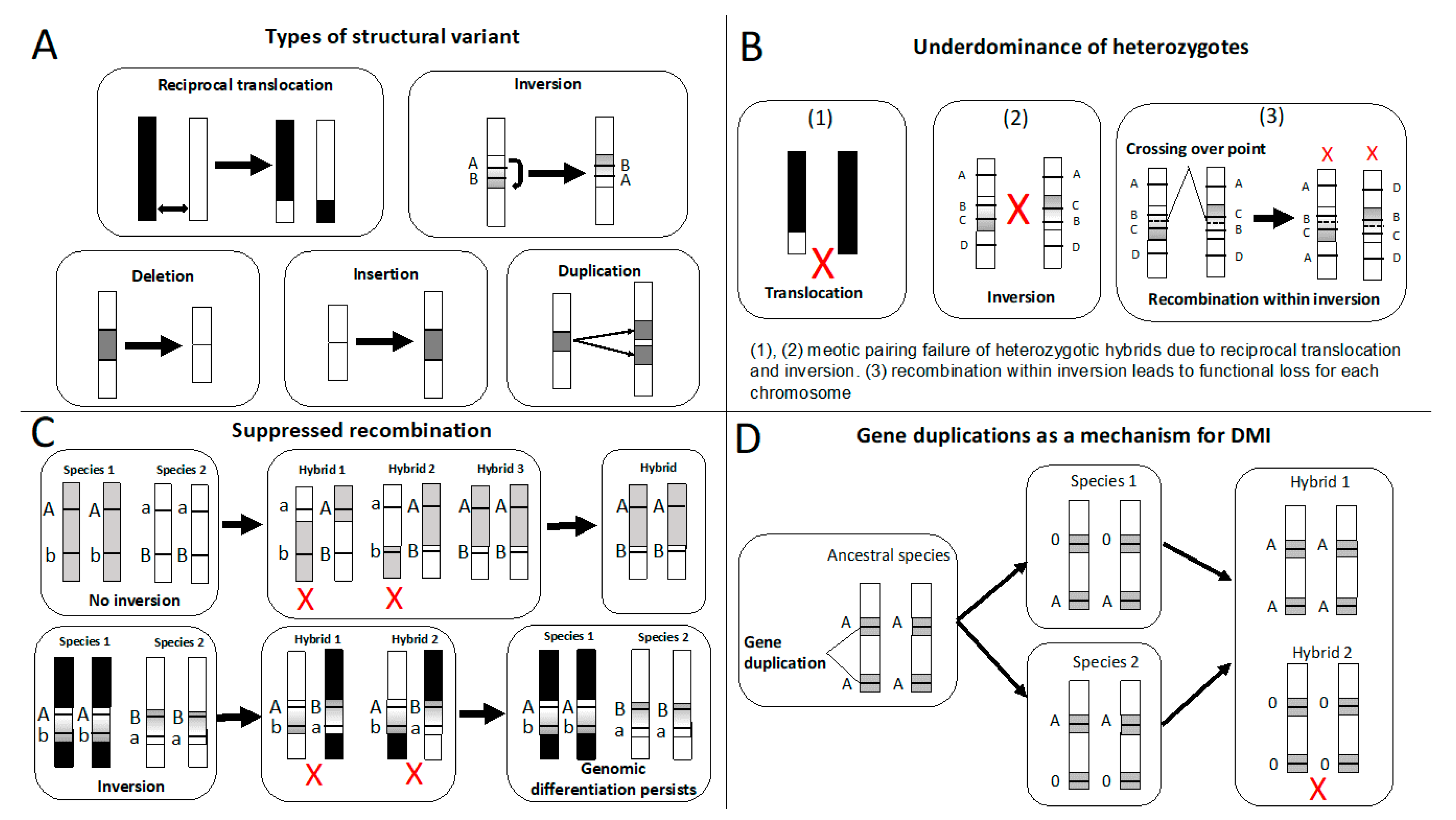
2. Reproductive Isolation Caused by Structural Variants: Theory and Evidence
2.1. Underdominance of Heterokaryotypes (Hybrid-Sterility Models)
2.2. Suppressed-Recombination Models
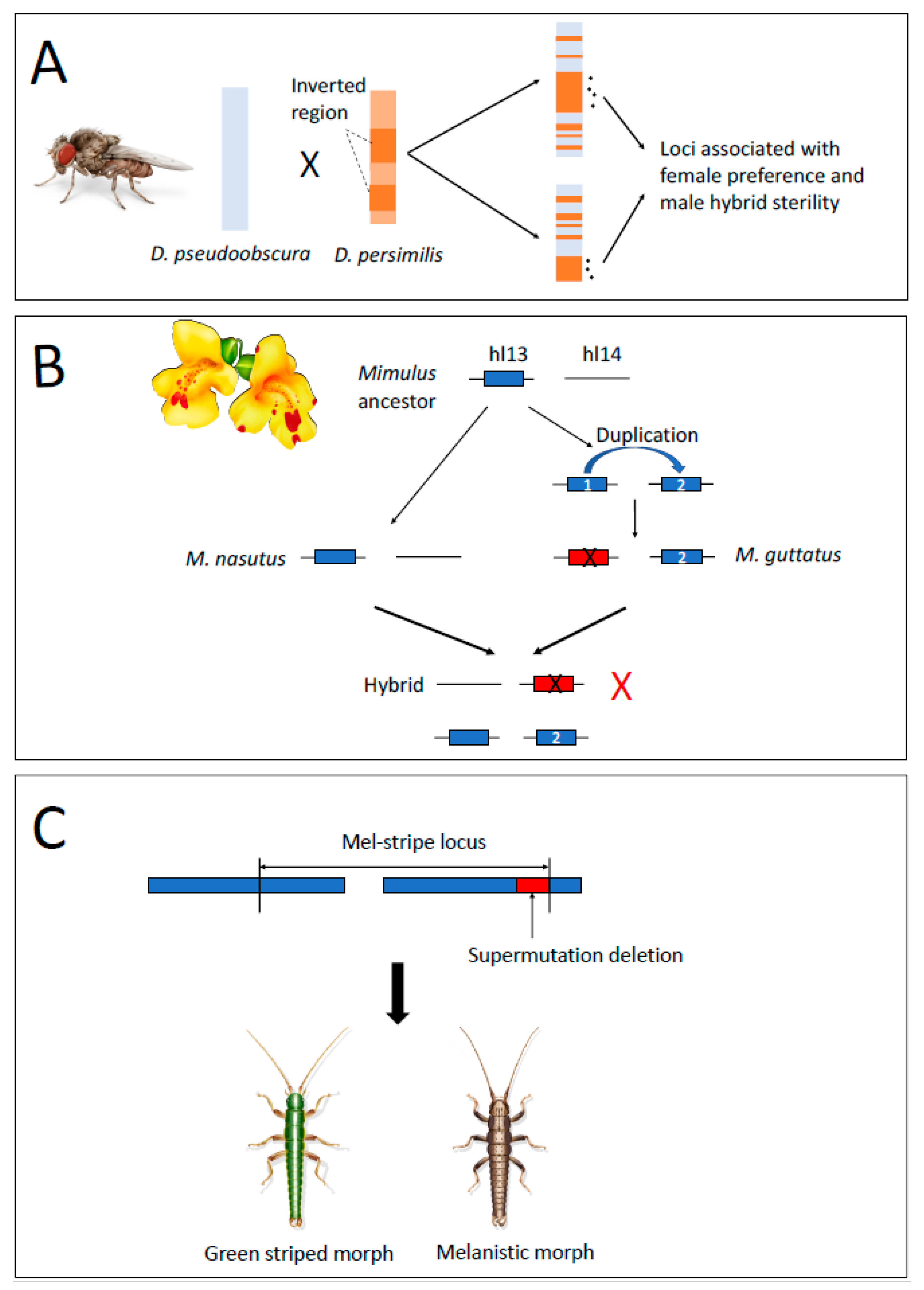
2.3. Gene Duplications as a Mechanism of Intrinsic Postzygotic Isolation
2.4. Structural Variants Act as Mutations of Large Effect Causing RI
3. Critical Knowledge Gaps and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, C.-I. The genic view of the process of speciation. J. Evol. Biol. 2001, 14, 851–865. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. Speciation; Sinauer: Sunderland, MA, USA, 2004; Volume 37, ISBN 9780878930890. [Google Scholar]
- Feder, J.L.; Xie, X.; Rull, J.; Velez, S.; Forbes, A.; Leung, B.; Dambroski, H.; Filchak, K.E.; Aluja, M. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc. Natl. Acad. Sci. USA 2005, 102, 6573–6580. [Google Scholar] [CrossRef] [PubMed]
- Kulmuni, J.; Butlin, R.K.; Lucek, K.; Savolainen, V.; Westram, A.M. Towards the completion of speciation: The evolution of reproductive isolation beyond the first barriers. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190528. [Google Scholar] [CrossRef] [PubMed]
- Ting, C.-T.; Tsaur, S.-C.; Sun, S.; Browne, W.; Chen, Y.-C.; Patel, N.; Wu, C.-I. Gene duplication and speciation in Drosophila: Evidence from the Odysseus locus. Proc. Natl. Acad. Sci. USA 2004, 101, 12232–12235. [Google Scholar] [CrossRef]
- Wittbrodt, J.; Adam, D.; Malitschek, B.; Mäueler, W.; Raulf, F.; Telling, A.; Robertson, S.M.; Schartl, M. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nat. Cell Biol. 1989, 341, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-I.; Ting, C.-T. Genes and speciation. Nat. Rev. Genet. 2004, 5, 114–122. [Google Scholar] [CrossRef]
- Orr, H.A.; Masly, J.; Phadnis, N. Speciation in Drosophila: From Phenotypes to Molecules. J. Hered. 2006, 98, 103–110. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Blackman, B. Speciation genes in plants. Ann. Bot. 2010, 106, 439–455. [Google Scholar] [CrossRef]
- Nosil, P.; Feder, J.L.; Gompert, Z. How many genetic changes create new species? Science 2021, 371, 777–779. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, E.R.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Fritz, M.H.-Y.; et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Catanach, A.; Crowhurst, R.; Deng, C.; David, C.; Bernatchez, L.; Wellenreuther, M. The genomic pool of standing structural variation outnumbers single nucleotide polymorphism by threefold in the marine teleost Chrysophrys auratus. Mol. Ecol. 2019, 28, 1210–1223. [Google Scholar] [CrossRef]
- Faria, R.; Chaube, P.; Morales, H.E.; Larsson, T.; Lemmon, A.R.; Lemmon, E.M.; Rafajlović, M.; Panova, M.; Ravinet, M.; Johannesson, K.; et al. Multiple chromosomal rearrangements in a hybrid zone between Littorina saxatilis ecotypes. Mol. Ecol. 2018, 28, 1375–1393. [Google Scholar] [CrossRef]
- Noor, M.A.F.; Grams, K.L.; Bertucci, L.A.; Reiland, J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 2001, 98, 12084–12088. [Google Scholar] [CrossRef]
- Sturtevant, A.H. A Case of Rearrangement of Genes in Drosophila. Proc. Natl. Acad. Sci. USA 1921, 7, 235–237. [Google Scholar] [CrossRef]
- Lewis, E.B. The Theory and Application of a New Method of Detecting Chromosomal Rearrangements in Drosophila melanogaster. Am. Nat. 1954, 88, 225–239. [Google Scholar] [CrossRef]
- White, M.; Key, K.; Andre, M.; Cheney, J. Cytogenetics of the Viatica group of morabine grasshoppers II.Kangaroo Island populations. Aust. J. Zoöl. 1969, 17, 313–328. [Google Scholar] [CrossRef]
- Creighton, H.B.; McClintock, B. A Correlation of Cytological and Genetical Crossing-Over in Zea Mays. Proc. Natl. Acad. Sci. USA 1931, 17, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Dobzhansky, T.G. Genetics of the Evolutionary Process; Columbia University Press: New York, NY, USA, 1971; ISBN 978-0-231-08306-5. [Google Scholar]
- Wallace, E.M. A possible case of affinity in tomatoes. Heredity 1960, 14, 275–283. [Google Scholar] [CrossRef]
- Lewis, H. Catastrophic Selection as a Factor in Speciation. Evolution 1962, 16, 257–271. [Google Scholar] [CrossRef]
- Zeitouni, B.; Boeva, V.; Janoueix-Lerosey, I.; Loeillet, S.; Legoix-Né, P.; Nicolas, A.; Delattre, O.; Barillot, E. SVDetect: A tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 2010, 26, 1895–1896. [Google Scholar] [CrossRef]
- Ma, J.; Amos, C.I. Investigation of Inversion Polymorphisms in the Human Genome Using Principal Components Analysis. PLoS ONE 2012, 7, e40224. [Google Scholar] [CrossRef]
- De Coster, W.; De Rijk, P.; De Roeck, A.; De Pooter, T.; D’Hert, S.; Strazisar, M.; Sleegers, K.; Van Broeckhoven, C. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 2019, 29, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Lucek, K.; Gompert, Z.; Nosil, P. The role of structural genomic variants in population differentiation and ecotype formation in Timema cristinae walking sticks. Mol. Ecol. 2019, 28, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Owens, G.L.; Bercovich, N.; Légaré, J.-S.; Soudi, S.; Burge, D.O.; Huang, K.; Ostevik, K.L.; Drummond, E.B.M.; Imerovski, I.; et al. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nat. Cell Biol. 2020, 584, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Ebert, P.; Audano, P.A.; Zhu, Q.; Rodriguez-Martin, B.; Porubsky, D.; Bonder, M.J.; Sulovari, A.; Ebler, J.; Zhou, W.; Mari, R.S.; et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 2021, 372, eabf7117. [Google Scholar] [CrossRef] [PubMed]
- Joron, M.; Frezal, L.; Jones, R.; Chamberlain, N.L.; Lee, S.F.; Haag, C.; Whibley, A.; Becuwe, M.; Baxter, S.W.; Ferguson, L.; et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nat. Cell Biol. 2011, 477, 203–206. [Google Scholar] [CrossRef]
- Jay, P.; Chouteau, M.; Whibley, A.; Bastide, H.; Parrinello, H.; Llaurens, V.; Joron, M. Mutation load at a mimicry supergene sheds new light on the evolution of inversion polymorphisms. Nat. Genet. 2021, 53, 288–293. [Google Scholar] [CrossRef]
- Villoutreix, R.; de Carvalho, C.F.; Soria-Carrasco, V.; Lindtke, D.; De-La-Mora, M.; Muschick, M.; Feder, J.L.; Parchman, T.L.; Gompert, Z.; Nosil, P. Large-scale mutation in the evolution of a gene complex for cryptic coloration. Science 2020, 369, 460–466. [Google Scholar] [CrossRef]
- Jones, F.; Broad Institute Genome Sequencing Platform; Whole Genome Assembly Team; Grabherr, M.G.; Chan, Y.F.; Russell, P.; Mauceli, E.; Johnson, J.; Swofford, R.; Pirun, M.; et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nat. Cell Biol. 2012, 484, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Nakazato, T.; Salmaso, M.; Burke, J.M.; Tang, S.; Knapp, S.J.; Rieseberg, L.H. Extensive Chromosomal Repatterning and the Evolution of Sterility Barriers in Hybrid Sunflower Species. Genetics 2005, 171, 291–303. [Google Scholar] [CrossRef]
- Hauffe, H.C.; Searle, J. Chromosomal Heterozygosity and Fertility in House Mice (Mus musculus domesticus) From Northern Italy. Genetics 1998, 150, 1143–1154. [Google Scholar] [CrossRef]
- Wang, R.L.; Hey, J. The Speciation History of Drosophila Pseudoobscura and Close Relatives: Inferences from DNA Sequence Variation at the Period Locus. Genetics 1996, 144, 1113–1126. [Google Scholar] [CrossRef]
- Piálek, J.; Hauffe, H.C.; Rodríguez-Clark, K.M.; Searle, J.B. Raciation and speciation in house mice from the Alps: The role of chromosomes. Mol. Ecol. 2008, 10, 613–625. [Google Scholar] [CrossRef]
- Garagna, S.; Page, J.; Fernandez-Donoso, R.; Zuccotti, M.; Searle, J.B. The Robertsonian phenomenon in the house mouse: Mutation, meiosis and speciation. Chromosoma 2014, 123, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Jolly, C.; Hoti, M.; Speed, D.; Shaw, L.; Rallis, C.; Balloux, F.; Dessimoz, C.; Bähler, J.; Sedlazeck, F.J. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017, 8, 14061. [Google Scholar] [CrossRef] [PubMed]
- Abril, V.V.; Carnelossi, E.A.G.; González, S.; Duarte, J.M.B. Elucidating the Evolution of the Red Brocket Deer Mazama Americana Complex (Artiodactyla; Cervidae). CGR 2010, 128, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Cursino, M.S.; Salviano, M.B.; Abril, V.V.; dos Santos Zanetti, E.; Duarte, J.M.B. The Role of Chromosome Variation in the Speciation of the Red Brocket Deer Complex: The Study of Reproductive Isolation in Females. BMC Evol. Biol. 2014, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Bracewell, R.R.; Bentz, B.J.; Sullivan, B.T.; Good, J.M. Rapid Neo-Sex Chromosome Evolution and Incipient Speciation in a Major Forest Pest. Nat. Commun. 2017, 8, 1593. [Google Scholar] [CrossRef] [PubMed]
- Dowle, E.J.; Bracewell, R.R.; Pfrender, M.E.; Mock, K.E.; Bentz, B.J.; Ragland, G.J. Reproductive Isolation and Environmental Adaptation Shape the Phylogeography of Mountain Pine Beetle (Dendroctonus Ponderosae). Mol. Ecol. 2017, 26, 6071–6084. [Google Scholar] [CrossRef]
- Ferree, P.M.; Barbash, D.A. Species-Specific Heterochromatin Prevents Mitotic Chromosome Segregation to Cause Hybrid Lethality in Drosophila. PLoS Biol. 2009, 7, e1000234. [Google Scholar] [CrossRef]
- Fishman, L.; Stathos, A.; Beardsley, P.M.; Williams, C.F.; Hill, J.P. Chromosomal Rearrangements and the genetics of reproductive barriers inmimulus(monkey flowerS). Evolution 2013, 67, 2547–2560. [Google Scholar] [CrossRef] [PubMed]
- Ayala, D.; Guerrero, R.F.; Kirkpatrick, M. Reproductive Isolation and Local Adaptation Quantified for a Chromosome Inversion in a Malaria Mosquito. Evolution 2013, 67, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Berdan, E.L.; Fuller, R.C.; Kozak, G.M. Genomic Landscape of Reproductive Isolation in Lucania Killifish: The Role of Sex Loci and Salinity. J. Evol. Biol. 2021, 34, 157–174. [Google Scholar] [CrossRef]
- Nosil, P.; Villoutreix, R.; de Carvalho, C.F.; Farkas, T.E.; Soria-Carrasco, V.; Feder, J.L.; Crespi, B.J.; Gompert, Z. Natural Selection and the Predictability of Evolution in Timema Stick Insects. Science 2018, 359, 765–770. [Google Scholar] [CrossRef]
- Hager, E.R.; Harringmeyer, O.S.; Wooldridge, T.B.; Theingi, S.; Gable, J.T.; McFadden, S.; Neugeboren, B.; Turner, K.M.; Hoekstra, H.E. A Chromosomal Inversion Drives Evolution of Multiple Adaptive Traits in Deer Mice. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lowry, D.B.; Willis, J.H. A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. PLoS Biol. 2010, 8, e1000500. [Google Scholar] [CrossRef]
- Oneal, E.; Lowry, D.B.; Wright, K.M.; Zhu, Z.; Willis, J.H. Divergent Population Structure and Climate Associations of a Chromosomal Inversion Polymorphism across the Mimulus Guttatus Species Complex. Mol. Ecol. 2014, 23, 2844–2860. [Google Scholar] [CrossRef]
- Twyford, A.D.; Friedman, J. Adaptive Divergence in the Monkey Flower Mimulus Guttatus Is Maintained by a Chromosomal Inversion: GENETICS OF ADAPTIVE DIVERGENCE IN MONKEY FLOWERS. Evolution 2015, 69, 1476–1486. [Google Scholar] [CrossRef]
- Feder, J.L.; Roethele, J.B.; Filchak, K.; Niedbalski, J.; Romero-Severson, J. Evidence for Inversion Polymorphism Related to Sympatric Host Race Formation in the Apple Maggot Fly, Rhagoletis Pomonella. Genetics 2003, 163, 939–953. [Google Scholar] [CrossRef]
- Manoukis, N.; Powell, J.R.; Toure, M.B.; Sacko, A.; Edillo, F.E.; Coulibaly, M.B.; Traore, S.F.; Taylor, C.E.; Besansky, N.J. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2008, 105, 2940–2945. [Google Scholar] [CrossRef] [PubMed]
- Kirubakaran, T.G.; Grove, H.; Kent, M.P.; Sandve, S.R.; Baranski, M.; Nome, T.; De Rosa, M.C.; Righino, B.; Johansen, T.; Otterå, H.; et al. Two adjacent inversions maintain genomic differentiation between migratory and stationary ecotypes of Atlantic cod. Mol. Ecol. 2016, 25, 2130–2143. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.M.; Griffith, S.C.; Price, T.D. Sex Chromosome Inversions Enforce Reproductive Isolation across an Avian Hybrid Zone. Mol. Ecol. 2019, 28, 1246–1262. [Google Scholar] [CrossRef]
- Huang, K.; Andrew, R.L.; Owens, G.L.; Ostevik, K.L.; Rieseberg, L.H. Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype. Mol. Ecol. 2020, 29, 2535–2549. [Google Scholar] [CrossRef]
- Lohse, K.; Clarke, M.; Ritchie, M.G.; Etges, W.J. Genome-Wide Tests for Introgression between Cactophilic Drosophila Implicate a Role of Inversions during Speciation. Evolution 2015, 69, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Zuellig, M.P.; Sweigart, A.L. Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS Genet. 2018, 14, e1007130. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Patel, D.; Le Metté, C.; Giorgi, V.; Camilleri, C.; Bennett, M.; Loudet, O. Divergent Evolution of Duplicate Genes Leads to Genetic Incompatibilities Within A. thaliana. Science 2009, 323, 623–626. [Google Scholar] [CrossRef]
- Masly, J.; Jones, C.D.; Noor, M.; Locke, J.; Orr, H.A. Gene Transposition as a Cause of Hybrid Sterility in Drosophila. Science 2006, 313, 1448–1450. [Google Scholar] [CrossRef] [PubMed]
- Weissensteiner, M.H.; Bunikis, I.; Catalán, A.; Francoijs, K.-J.; Knief, U.; Heim, W.; Peona, V.; Pophaly, S.D.; Sedlazeck, F.; Suh, A.; et al. Discovery and population genomics of structural variation in a songbird genus. Nat. Commun. 2020, 11, 3403. [Google Scholar] [CrossRef]
- North, H.L.; Caminade, P.; Severac, D.; Belkhir, K.; Smadja, C.M. The Role of Copy-Number Variation in the Reinforcement of Sexual Isolation between the Two European Subspecies of the House Mouse. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190540. [Google Scholar] [CrossRef]
- Michael, J.D.; White, W.H. VIII + 456 pp. illus. $27.50. Paleobiology 1978, 4, 373–379. [Google Scholar] [CrossRef]
- Barton, N.; Bengtsson, B.O. The barrier to genetic exchange between hybridising populations. Heredity 1986, 57, 357–376. [Google Scholar] [CrossRef] [PubMed]
- Coyne, J.A.; Meyers, W.; Crittenden, A.P.; Sniegowski, P. The fertility effects of pericentric inversions in Drosophila mela-nogaster. Genetics 1993, 134, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Homolka, D.; Ivánek, R.; Capkova, J.; Jansa, P.; Forejt, J. Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res. 2007, 17, 1431–1437. [Google Scholar] [CrossRef] [PubMed]
- Delneri, D.; Colson, I.; Grammenoudi, S.; Roberts, I.N.; Louis, E.; Oliver, S.G. Engineering evolution to study speciation in yeasts. Nat. Cell Biol. 2003, 422, 68–72. [Google Scholar] [CrossRef]
- Rieseberg, L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001, 16, 351–358. [Google Scholar] [CrossRef]
- Orr, H.A.; Coyne, J.A. The genetics of postzygotic isolation in the Drosophila virilis group. Genetics 1989, 121, 527–537. [Google Scholar] [CrossRef]
- Divina, P.; Storchová, R.; Gregorová, S.; Buckiová, D.; Kyselová, V.; Forejt, J. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 2004, 15, 515–524. [Google Scholar] [CrossRef]
- Masly, J.P.; Presgraves, D.C. High-Resolution Genome-Wide Dissection of the Two Rules of Speciation in Drosophila. PLoS Biol. 2007, 5, e243. [Google Scholar] [CrossRef]
- Good, J.M.; Dean, M.D.; Nachman, M.W. A Complex Genetic Basis to X-Linked Hybrid Male Sterility Between Two Species of House Mice. Genetics 2008, 179, 2213–2228. [Google Scholar] [CrossRef]
- Matsuda, Y.; Hirobe, T.; Chapman, V.M. Genetic basis of X-Y chromosome dissociation and male sterility in interspecific hybrids. Proc. Natl. Acad. Sci. USA 1991, 88, 4850–4854. [Google Scholar] [CrossRef]
- Presgraves, D.C. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008, 24, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Presgraves, D.C. Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol. Ecol. 2018, 27, 3822–3830. [Google Scholar] [CrossRef]
- Janoušek, V.; Fischerová, J.; Mořkovský, L.; Reif, J.; Antczak, M.; Albrecht, T.; Reifová, R. Postcopulatory sexual selection reduces Z-linked genetic variation and might contribute to the large Z effect in passerine birds. Heredity 2019, 122, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Lande, R. Effective Deme Sizes During Long-Term Evolution Estimated from Rates of Chromosomal Rearrangement. Evolution 1979, 33, 234. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H. The fitness of hybrids between two chromosomal races of the grasshopper Podisma pedestris. Heredity 1980, 45, 47–59. [Google Scholar] [CrossRef]
- Baker, R.J.; Bickham, J.W. Speciation by monobrachial centric fusions. Proc. Natl. Acad. Sci. USA 1986, 83, 8245–8248. [Google Scholar] [CrossRef]
- Nachman, M.W.; Searle, J. Why is the house mouse karyotype so variable? Trends Ecol. Evol. 1995, 10, 397–402. [Google Scholar] [CrossRef]
- Navarro, A.; Barton, N.H. Accumulating postzygotic isolation genes in parapatry: A new twist on chromosomal speciation. Evolution 2003, 57, 447–459. [Google Scholar] [CrossRef]
- Navarro, A.; Betran, E.; Barbadilla, A.; Ruiz, A. Recombination and Gene Flux Caused by Gene Conversion and Crossing Over in Inversion Heterokaryotypes. Genetics 1997, 146, 695–709. [Google Scholar] [CrossRef]
- Butlin, R.K. Recombination and speciation. Mol. Ecol. 2005, 14, 2621–2635. [Google Scholar] [CrossRef]
- Rieseberg, L.; Whitton, J.; Gardner, K. Hybrid Zones and the Genetic Architecture of a Barrier to Gene Flow between Two Sunflower Species. Genetics 1999, 152, 713–727. [Google Scholar] [CrossRef]
- Machado, C.A.; Kliman, R.M.; Markert, J.A.; Hey, J. Inferring the history of speciation from multilocus DNA sequence data: The case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 2002, 19, 472–488. [Google Scholar] [CrossRef] [PubMed]
- Noor, M.A.F.; Garfield, D.A.; Schaeffer, S.; Machado, C.A. Divergence between the Drosophila pseudoobscura and D. persimilis Genome Sequences in Relation to Chromosomal Inversions. Genetics 2007, 177, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Kulathinal, R.J.; Stevison, L.; Noor, M.A.F. The Genomics of Speciation in Drosophila: Diversity, Divergence, and Introgression Estimated Using Low-Coverage Genome Sequencing. PLoS Genet. 2009, 5, e1000550. [Google Scholar] [CrossRef] [PubMed]
- Stevison, L.S.; Hoehn, K.B.; Noor, M. Effects of Inversions on Within- and Between-Species Recombination and Divergence. Genome Biol. Evol. 2011, 3, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wurm, Y.; Nipitwattanaphon, M.; Riba-Grognuz, O.; Huang, Y.-C.; Shoemaker, D.; Keller, L. A Y-like social chromosome causes alternative colony organization in fire ants. Nat. Cell Biol. 2013, 493, 664–668. [Google Scholar] [CrossRef]
- Korunes, K.L.; Noor, M.A.F. Pervasive gene conversion in chromosomal inversion heterozygotes. Mol. Ecol. 2018, 28, 1302–1315. [Google Scholar] [CrossRef]
- Kirkpatrick, M.; Barton, N. Chromosome Inversions, Local Adaptation and Speciation. Genetics 2005, 173, 419–434. [Google Scholar] [CrossRef]
- Charlesworth, B.; Barton, N.H. The Spread of an Inversion with Migration and Selection. Genetics 2018, 208, 377–382. [Google Scholar] [CrossRef]
- Berg, P.R.; Star, B.; Pampoulie, C.; Sodeland, M.; Barth, J.; Knutsen, H.; Jakobsen, K.S.; Jentoft, S. Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Sci. Rep. 2016, 6, 23246. [Google Scholar] [CrossRef]
- Michel, A.P.; Sim, S.; Powell, T.H.Q.; Taylor, M.S.; Nosil, P.; Feder, J.L. Widespread genomic divergence during sympatric speciation. Proc. Natl. Acad. Sci. USA 2010, 107, 9724–9729. [Google Scholar] [CrossRef]
- Hooper, D.M.; Price, T.D. Chromosomal inversion differences correlate with range overlap in passerine birds. Nat. Ecol. Evol. 2017, 1, 1526–1534. [Google Scholar] [CrossRef]
- Brown, K.M.; Burk, L.M.; Henagan, L.M.; Noor, M.A.F. A test of the chromosomal rearrangement model of speciation in drosophila pseudoobscura. Evolution 2004, 58, 1856–1860. [Google Scholar] [CrossRef]
- Kozak, G.M.; Wadsworth, C.B.; Kahne, S.; Bogdanowicz, S.M.; Harrison, R.G.; Coates, B.; Dopman, E. A combination of sexual and ecological divergence contributes to rearrangement spread during initial stages of speciation. Mol. Ecol. 2017, 26, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-R.; Wang, B.; Mojica, J.P.; Mandáková, T.; Prasad, K.V.S.K.; Goicoechea, J.L.; Perera, N.; Hellsten, U.; Hundley, H.N.; Johnson, J.; et al. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat. Ecol. Evol. 2017, 1, 1–13. [Google Scholar] [CrossRef]
- Coughlan, J.M.; Willis, J.H. Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex. Mol. Ecol. 2018, 28, 1343–1357. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Barker, S.L.; Rastas, P.M.; Pinharanda, A.; Martin, S.H.; Durbin, R.; McMillan, W.O.; Merrill, R.M.; Jiggins, C.D. No evidence for maintenance of a sympatric Heliconius species barrier by chromosomal inversions. Evol. Lett. 2017, 1, 138–154. [Google Scholar] [CrossRef]
- Feder, J.L.; Nosil, P. Chromosomal Inversions and species differences: When are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution 2009, 63, 3061–3075. [Google Scholar] [CrossRef] [PubMed]
- Crown, K.N.; Miller, D.E.; Sekelsky, J.; Hawley, R.S. Local Inversion Heterozygosity Alters Recombination throughout the Genome. Curr. Biol. 2018, 28, 2984–2990.e3. [Google Scholar] [CrossRef]
- Haldane, J.B.S. The Part Played by Recurrent Mutation in Evolution. Am. Nat. 1933, 67, 5–19. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.-L.; Postlethwait, J. Preservation of Duplicate Genes by Complementary, Degenerative Mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef]
- Lynch, M.; Force, A. The Probability of Duplicate Gene Preservation by Subfunctionalization. Genetics 2000, 154, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; O’Hely, M.; Walsh, B.; Force, A. The Probability of Preservation of a Newly Arisen Gene Duplicate. Genetics 2001, 159, 1789–1804. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, J.H.; Sankoff, D. Comparable Rates of Gene Loss and Functional Divergence After Genome Duplications Early in Vertebrate Evolution. Genetics 1997, 147, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.H.; Yan, Y.-L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z.; et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef]
- Duvaux, L.; Geissmann, Q.; Gharbi, K.; Zhou, J.-J.; Ferrari, J.; Smadja, C.M.; Butlin, R.K. Dynamics of copy number variation in host races of the pea aphid. Mol. Biol. Evol. 2014, 32, 63–80. [Google Scholar] [CrossRef]
- Mizuta, Y.; Harushima, Y.; Kurata, N. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 2010, 107, 20417–20422. [Google Scholar] [CrossRef]
- Kirkpatrick, M. How and Why Chromosome Inversions Evolve. PLoS Biol. 2010, 8, e1000501. [Google Scholar] [CrossRef]
- Guerrero, R.F.; Rousset, F.; Kirkpatrick, M. Coalescent patterns for chromosomal inversions in divergent populations. Philos. Trans. R. Soc. B Biol. Science 2012, 367, 430–438. [Google Scholar] [CrossRef]
- Hof, A.E.V.; Campagne, P.; Rigden, D.; Yung, C.J.; Lingley, J.; Quail, M.A.; Hall, N.; Darby, A.; Saccheri, I.J. The industrial melanism mutation in British peppered moths is a transposable element. Nat. Cell Biol. 2016, 534, 102–105. [Google Scholar] [CrossRef]
- Chan, Y.F.; Marks, M.E.; Jones, F.; Villarreal, G.; Shapiro, M.D.; Brady, S.D.; Southwick, A.M.; Absher, D.M.; Grimwood, J.; Schmutz, J.; et al. Adaptive Evolution of Pelvic Reduction in Sticklebacks by Recurrent Deletion of a Pitx1 Enhancer. Science 2009, 327, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Comeault, A.A.; Flaxman, S.M.; Riesch, R.; Curran, E.; Soria-Carrasco, V.; Gompert, Z.; Farkas, T.E.; Muschick, M.; Parchman, T.L.; Schwander, T.; et al. Selection on a Genetic Polymorphism Counteracts Ecological Speciation in a Stick Insect. Curr. Biol. 2015, 25, 1975–1981. [Google Scholar] [CrossRef]
- Rundle, H.D.; Nosil, P. Ecological speciation. Ecol. Lett. 2005, 8, 336–352. [Google Scholar] [CrossRef]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Futuyma, D.J.; Shapiro, L.H.; Harrison, R.G. Hybrid Zones and the Evolutionary Process. Evolution 1995, 49, 222. [Google Scholar] [CrossRef]
- Gompert, Z.; Buerkle, C.A. A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Mol. Ecol. 2009, 18, 1207–1224. [Google Scholar] [CrossRef]
- Payseur, B.A.; Rieseberg, L.H. A genomic perspective on hybridization and speciation. Mol. Ecol. 2016, 25, 2337–2360. [Google Scholar] [CrossRef] [PubMed]
- Gompert, Z.; Mandeville, E.G.; Buerkle, C.A. Analysis of Population Genomic Data from Hybrid Zones. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 207–229. [Google Scholar] [CrossRef]
- Mahmoud, M.; Gobet, N.; Cruz-Dávalos, D.I.; Mounier, N.; Dessimoz, C.; Sedlazeck, F.J. Structural variant calling: The long and the short of it. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Lindtke, D.; Yeaman, S. Identifying the loci of speciation: The challenge beyond genome scans. J. Evol. Biol. 2017, 30, 1478–1481. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Q.; Wang, Q.; Jia, P.; Zhao, Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: Features and perspectives. BMC Bioinform. 2013, 14, S1. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.; Wang, H.; Kang, H.; Zhang, Q.; Liu, J.-F. Enhancing Genome-Wide Copy Number Variation Identification by High Density Array CGH Using Diverse Resources of Pig Breeds. PLoS ONE 2014, 9, e87571. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate Detection of Complex Structural Variations Using Single Molecule Sequencing. Nat Methods 2018, 15, 461–468. [Google Scholar] [CrossRef]
- Heller, D.; Vingron, M. SVIM: Structural Variant Identification Using Mapped Long Reads. Bioinformatics 2019, 35, 2907–2915. [Google Scholar] [CrossRef]
- Kiełbasa, S.M.; Wan, R.; Sato, K.; Horton, P.; Frith, M.C. Adaptive Seeds Tame Genomic Sequence Comparison. Genome Res. 2011, 21, 487–493. [Google Scholar] [CrossRef]
- Tham, C.Y.; Tirado-Magallanes, R.; Goh, Y.; Fullwood, M.J.; Koh, B.T.H.; Wang, W.; Ng, C.H.; Chng, W.J.; Thiery, A.; Tenen, D.G.; et al. NanoVar: Accurate Characterization of Patients’ Genomic Structural Variants Using Low-Depth Nanopore Sequencing. Genome Biol. 2020, 21, 56. [Google Scholar] [CrossRef]
- Cretu Stancu, M.; van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; de Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Espejo Valle-Inclan, J.; et al. Mapping and Phasing of Structural Variation in Patient Genomes Using Nanopore Sequencing. Nat. Commun. 2017, 8, 1326. [Google Scholar] [CrossRef]
- Chaisson, M.J.; Tesler, G. Mapping Single Molecule Sequencing Reads Using Basic Local Alignment with Successive Refinement (BLASR): Application and Theory. BMC Bioinform. 2012, 13, 238. [Google Scholar] [CrossRef]
- English, A.C.; Salerno, W.J.; Reid, J.G. PBHoney: Identifying Genomic Variants via Long-Read Discordance and Interrupted Mapping. BMC Bioinform. 2014, 15, 180. [Google Scholar] [CrossRef]
- Huddleston, J.; Chaisson, M.J.P.; Steinberg, K.M.; Warren, W.; Hoekzema, K.; Gordon, D.; Graves-Lindsay, T.A.; Munson, K.M.; Kronenberg, Z.N.; Vives, L.; et al. Discovery and Genotyping of Structural Variation from Long-Read Haploid Genome Sequence Data. Genome Res. 2017, 27, 677–685. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, Y.; Jiang, Y.; Li, J.; Gao, Y.; Cui, Z.; Liu, Y.; Liu, B.; Wang, Y. Long-Read-Based Human Genomic Structural Variation Detection with CuteSV. Genome Biol. 2020, 21, 189. [Google Scholar] [CrossRef] [PubMed]
- PacificBiosciences/Pbmm2; Pacific Biosciences of California, Inc.: Menlo Park, CA, USA, 2021.
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef] [PubMed]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A Pattern Growth Approach to Detect Break Points of Large Deletions and Medium Sized Insertions from Paired-End Short Reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ruan, J.; Durbin, R. Mapping Short DNA Sequencing Reads and Calling Variants Using Mapping Quality Scores. Genome Res. 2008, 18, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- NovoAlign; Novocraft Technologies Sdn Bhd: Selangor, Malaysia, 2021.
- Homer, N.; Merriman, B.; Nelson, S.F. BFAST: An Alignment Tool for Large Scale Genome Resequencing. PLoS ONE 2009, 4, e7767. [Google Scholar] [CrossRef]
- Chen, K.; Wallis, J.W.; McLellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; McGrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An Algorithm for High-Resolution Mapping of Genomic Structural Variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef]
- Lunter, G.; Goodson, M. Stampy: A Statistical Algorithm for Sensitive and Fast Mapping of Illumina Sequence Reads. Genome Res. 2011, 21, 936–939. [Google Scholar] [CrossRef]
- SMALT—Wellcome Sanger Institute. Available online: https://www.sanger.ac.uk/tool/smalt-0/ (accessed on 16 July 2021).
- Gan, X.; Stegle, O.; Behr, J.; Steffen, J.G.; Drewe, P.; Hildebrand, K.L.; Lyngsoe, R.; Schultheiss, S.J.; Osborne, E.J.; Sreedharan, V.T.; et al. Multiple Reference Genomes and Transcriptomes for Arabidopsis Thaliana. Nature 2011, 477, 419–423. [Google Scholar] [CrossRef]
- Rimmer, A.; Phan, H.; Mathieson, I.; Iqbal, Z.; Twigg, S.R.F.; WGS500 Consortium; Wilkie, A.O.M.; McVean, G.; Lunter, G. Integrating Mapping-, Assembly- and Haplotype-Based Approaches for Calling Variants in Clinical Sequencing Applications. Nat. Genet. 2014, 46, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, D.P.; Tyukmaeva, V.I.; Gompert, Z.; Feder, J.; Nosil, P. Functional Genomics Offers New Tests of Speciation Hypotheses. Trends Ecol. Evol. 2020, 35, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Bush, G.L.; Case, S.M.; Wilson, A.C.; Patton, J.L. Rapid speciation and chromosomal evolution in mammals. Proc. Natl. Acad. Sci. USA 1977, 74, 3942–3946. [Google Scholar] [CrossRef]
- Leaché, A.D.; Banbury, B.L.; Linkem, C.W.; De Oca, A.N.-M. Phylogenomics of a rapid radiation: Is chromosomal evolution linked to increased diversification in north american spiny lizards (Genus Sceloporus)? BMC Evol. Biol. 2016, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- De Vos, J.M.; Augustijnen, H.; Bätscher, L.; Lucek, K. Speciation through chromosomal fusion and fission in Lepidoptera. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190539. [Google Scholar] [CrossRef]
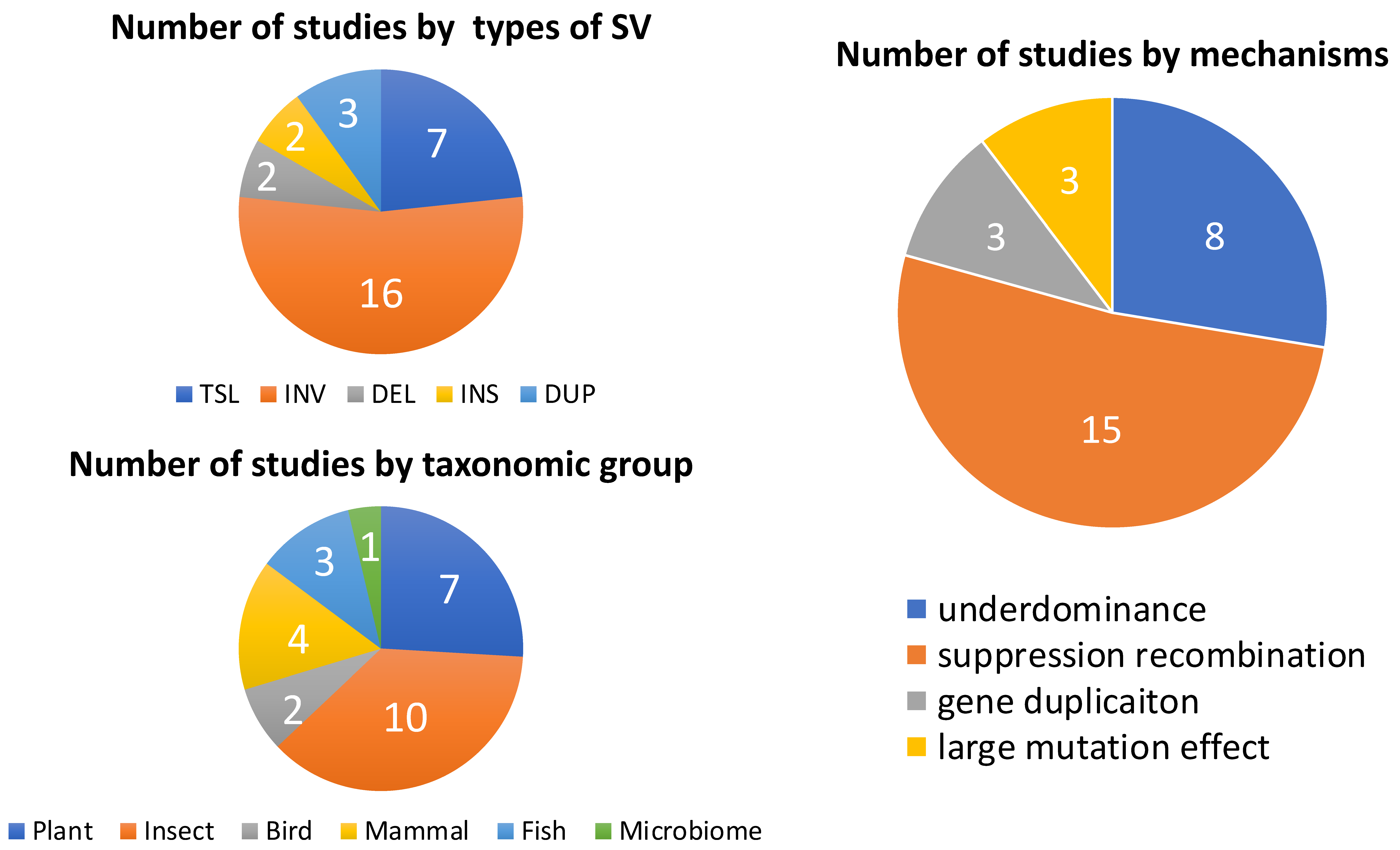
| Mechanism by Which SV Contributes to RI | SV Type | Study Organism | Affected RI | References |
|---|---|---|---|---|
| Underdominance | TSL | Sunflower hybrid species and their parental species (H. annuus and H. petiolaris) | Pollen sterility in hybrids. | Lai et al., 2005 [32] |
| Underdominance | TSL | House mouse Mus musculus domesticus | Hybrid subfertility and sterility | Hauffe and Searle 1993 [33]; Nachman and Searle 1995 [34]; Pialek et al., 2008 [35]; Garagna et al., 2014 [36] |
| Underdominance | TSL, INV | Fission yeast Schizosaccharomyces pombe | Intrinsic hybrid inviability | Jeffares et al., 2017 [37] |
| Underdominance | TSL, INV | Red brocket deer Mazama americana | Hybrid subfertility and sterility | Abril et al., 2010 [38], Cursino et al., 2014 [39] |
| Underdominance | DEL | Mountain pine beetle Dendroctonus ponderosae | Hybrid male sterility | Bracewell et al., 2017 [40], Dowle et al., 2017 [41] |
| Underdominance | INS | Fruit flies Drosophila simulans and D. melanogaster | Hybrid lethality | Ferree and Barbash 2009 [42] |
| Suppressed recombination and underdominance of heterokaryotypes | INV, TSL | Monkeyflowers Mimulus cardinalis and M. lewisii | Ecological isolation caused by adaptation to different elevation ranges and pollinators. Hybrid sterility. | Fishman et al., 2013 [43] |
| Suppressed recombination and underdominance of heterokaryons | INV | Mosquito Anopheles funestus | Assortative mating and postzygotic isolation resulting from underdominance of heterozygotes in particular habitats. | Ayala, Guerrero and Kirkpatrick 2013 [44] |
| Suppressed recombination and possibly underdominance of heterokaryons | TSL | Killifish Lucania goodei and L. parva | Hybrid inviability, behavioral isolation | Berdan, Fuller and Kozak 2021 [45] |
| Suppressed recombination | INV | Stick insect Timema cristinae | Habitat isolation associated with different cryptic color patterns (specialization to different host plants). | Nosil et al., 2018 [46], Lucek et al., 2019 [25] |
| Suppressed recombination | INV | Deer mouse Peromyscus maniculatus | Ecological isolation (local adaptation to different environments). | Hager et al., 2021 [47] |
| Suppressed recombination | INV | Three-spined stickleback (Gasterosteus aculeatus) | Ecological isolation caused by adaptation to freshwater and marine environment. | Jones et al., 2012 [31] |
| Suppressed recombination | INV | Yellow monkeyflower Mimulus guttatus | Ecological isolation including temporal isolation and habitat isolation. | Lowry and Willis 2010 [48], Oneal et al., 2014 [49], Twyrord and Friedman 2015 [50] |
| Suppressed recombination | INV | Apple maggot fly Rhagoletis pomonella | Ecological isolation including temporal isolation caused by adaptation to different host plants. | Feder et al., 2003 [51] |
| Suppressed recombination | INV | Fruit flies Drosophila pseudoobscura and D. persimilis | F1 hybrid male sterility, backcross hybrid inviability, F1 hybrid male courtship dysfunction, female species-specific preferences | Noor et al., 2001 [14] |
| Suppressed recombination | INV | Mosquito Anopheles gambiae | Ecological isolation caused by divergence in breeding sites. | Manoukis et al., 2008 [52] |
| Suppressed recombination | INV | The Northeast Arctic cod and Norwegian coastal cod populations of the Atlantic cod | Ecological isolation caused by different migration behaviors. | Kirubakaran et al., 2016 [53] |
| Suppressed recombination | INV | Long-tailed finch Poephila acuticauda | Unknown. Potentially prezygotic isolation caused by differences in bill color or sperm morphology between species. | Hooper et al., 2019 [54] |
| Suppressed recombination | INV | Prairie sunflower Helianthus petiolaris | Ecological isolation caused by adaptation to different environments | Huang et al., 2020 [55] |
| Suppressed recombination | INV | Fruit flies Drosophila mojavensis and D. arizonae | Unclear | Lohse et al., 2015 [56] |
| Suppressed recombination | INV | Sunflower species Helianthus annuus, H. petiolaris and H. argophyllus | Ecological isolation including temporal isolation caused by different flowering times | Todesco et al., 2020 [26] |
| Change of gene position caused by gene duplication and functional loss of ancestral copy in one species. | DUP | Monkeyflowers Mimulus guttatus and M. nasutus | F2 hybrid inviability | Zuellig and Sweigart 2018 [57] |
| Ancestral gene duplication has predisposed one of the paralogs for fast evolutionary rates. | DUP | Fruit flies Drosophila mauritiana and D. simulans | Hybrid male sterility | Ting et al 2004 [5] |
| Reciprocal gene loss after duplication | DUP | Wild strains of the plant Arabidopsis thaliana | Hybrid lethality in F2 cross | Bikard et al., 2009 [58] |
| Change of gene position | TSL | Fruit flies Drosophila melanogaster and D. simulans | F2 hybrid sterility | Masly et al., 2006 [59] |
| Mutations induced by SV | INS | Crow subspecies Corvus corone corone and C. c. cornix | Premating isolation | Weissensteiner et al., 2020 [60] |
| Source of mutations | DEL | House mouse Mus musculus musculus and M. m. domesticus | Assortative mating in the secondary contact zone (reinforcement) | North et al., 2020 [61] |
| Sequencing Method | Sequencing Platform | Alignment Method (Software) | Variant Calling (Software) | SV Types | Author (Study) |
|---|---|---|---|---|---|
| Long-read sequencing | ONT, PacBio | BWA-MEM [125], Minimap2 [126], NGMLR [127] | Sniffles | DEL, DUP, INS, INV, TRA, nested SVs (INVDUP, INVDEL) | Sedlazeck et al., 2018 [127] |
| ONT, PacBio | Minimap2 [126], NGMLR [126] | SVIM | DEL, DUP, INS, INV, TRA | Heller and Vingron, 2019 [128] | |
| ONT, PacBio | Minimap2 [126], LAST [129] | NanoVar | DEL, INV, DUP, INS, TRA | Tham et al., 2020 [130] | |
| ONT, PacBio | BWA-MEM [125], Minimap2 [126], NGMLR [127], LAST [129] | NanoSV | DEL, INS, DUP, INV, TRA | Stancu et al., 2017 [131] | |
| PacBio | BLASR [132] | PBHoney | INS, DEL, INV, TRA | English et al., 2014 [133] | |
| PacBio | BLASR [132] | SMRT-SV | INS, DEL, INV | Huddleston et al., 2017 [134] | |
| ONT, PacBio | Minimap2 [126] | cuteSV | DEL, INS, DUP, INV, TRA | Jiang et al., 2020 [135] | |
| PacBio | PBMM2 [136] | PBSV | INS, DEL, INV, DUP, TRA | ||
| Short-read sequencing | Illumina (short read sequencing platforms in general) | BWA-MEM [125] | LUMPY | DEL, DUP, INV, TRA | Layer et al., 2014 [137] |
| Illumina (short read sequencing platforms in general) | BWA-MEM [125] | DELLY | DEL, DUP, INV, TRA | Rausch et al., 2012 [138] | |
| Illumina (short read sequencing platforms in general) | BWA-MEM [125] | Pindel | DEL, INS, INV, DUP, TRA | Ye et al., 2009 [139] | |
| Illumina (short read sequencing platforms in general)—paired-end sequencing reads | MAQ [140], BWA [141], NovoAlign [142], Bfast [143] | BreakDancer | DEL, INS, INV, TRA | Chen et al., 2009 [144] | |
| Illumina | Stampy [145], BWA [141], SMALT [146], MAQ [140] | IMR/DENOM | INS, DEL | Gan et al., 2011 [147] | |
| Illumina | Stampy [145], BWA [141] | Platypus | INS, DEL | Rimmer et al., 2014 [148] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Reifová, R.; Halenková, Z.; Gompert, Z. How Important Are Structural Variants for Speciation? Genes 2021, 12, 1084. https://doi.org/10.3390/genes12071084
Zhang L, Reifová R, Halenková Z, Gompert Z. How Important Are Structural Variants for Speciation? Genes. 2021; 12(7):1084. https://doi.org/10.3390/genes12071084
Chicago/Turabian StyleZhang, Linyi, Radka Reifová, Zuzana Halenková, and Zachariah Gompert. 2021. "How Important Are Structural Variants for Speciation?" Genes 12, no. 7: 1084. https://doi.org/10.3390/genes12071084
APA StyleZhang, L., Reifová, R., Halenková, Z., & Gompert, Z. (2021). How Important Are Structural Variants for Speciation? Genes, 12(7), 1084. https://doi.org/10.3390/genes12071084






