CircVAMP3: A circRNA with a Role in Alveolar Rhabdomyosarcoma Cell Cycle Progression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Transfections
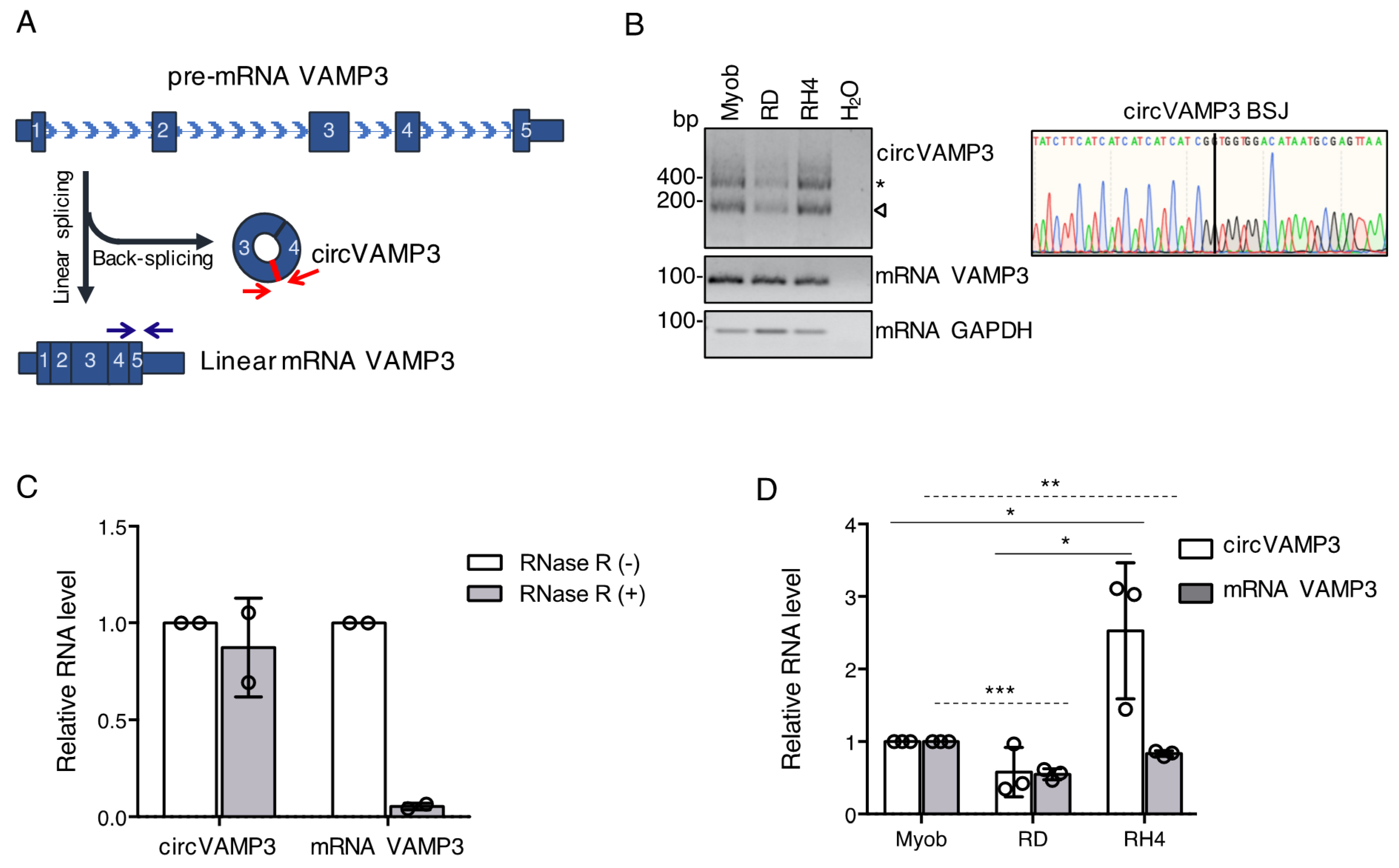
2.2. RNA Isolation and RNase-R Treatment
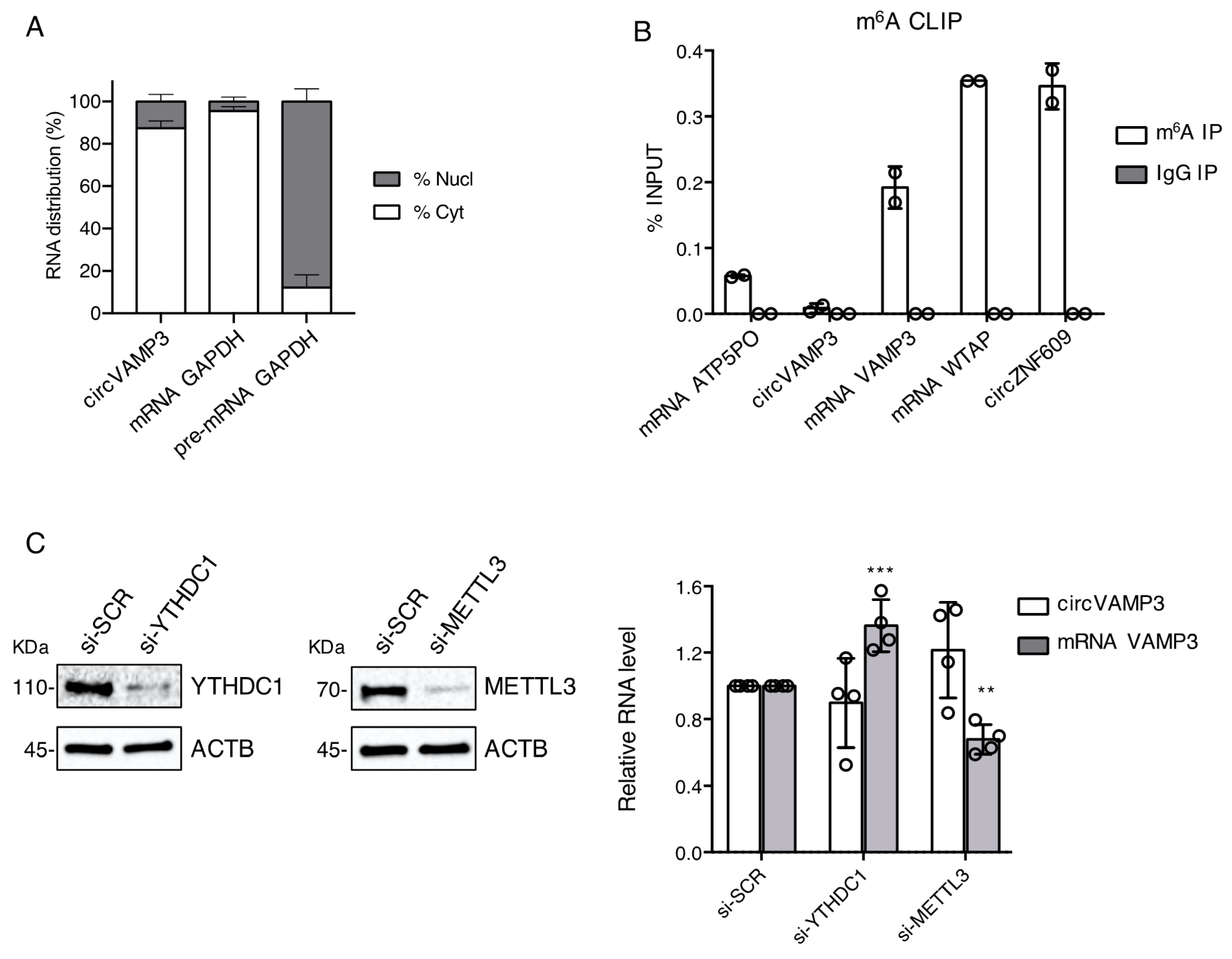
2.3. Nucleus-Cytoplasm Subcellular Fractionation
2.4. Western Blot
2.5. M6A CLIP
2.6. Flow Cytometric Analysis of Cell Cycle
2.7. RNA-Sequencing and Bioinformatic Analyses
3. Results
3.1. CircVAMP3 Is Highly Expressed in RH4 Cells
3.2. CircVAMP3 Is Located in the Cytoplasm and Has a Differential m6A Methylation with Respect to Its Linear Counterpart
3.3. CircVAMP3 Knock-Down Affects Proliferation-Related Pathways in ARMS Cells
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, 90859. [Google Scholar] [CrossRef] [Green Version]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; Herzog, M.; et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2014, 58, 870–885. [Google Scholar] [CrossRef] [Green Version]
- Di Timoteo, G.; Rossi, F.; Bozzoni, I. Circular RNAs in cell differentiation and development. Development 2020, 147. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant CircHIPK3 that regulates cell growth by sponging multiple MiRNAs. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes MiRNA deregulation and affects brain function. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with P21 and CDK2. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66. [Google Scholar] [CrossRef] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Begum, S.; Yiu, A.; Stebbing, J.; Castellano, L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene 2018, 37, 4055–4057. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Bach, D.H.; Lee, S.K.; Sood, A.K. Circular RNAs in cancer. Mol. Ther. Nucleic Acids 2019, 16, 118–129. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Wang, Y.; Fan, Y.; Fang, N.; Wang, T.; Xu, T.; Shu, Y. CircRNAs in cancer metabolism: A review. J. Hematol. Oncol. 2019, 12, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic role of fusion-CircRNAs derived from cancer-associated chromosomal translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [Green Version]
- Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of CircRNA metabolism by M6A modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef]
- Huh, W.; Egas Bejar, D. Rhabdomyosarcoma in adolescent and young adult patients: Current perspectives. Adolesc. Health Med. Ther. 2014, 5, 115. [Google Scholar] [CrossRef] [Green Version]
- Soleimani, V.D.; Rudnicki, M.A. New insights into the origin and the genetic basis of rhabdomyosarcomas. Cancer Cell 2011, 19, 157–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashi, V.P.; Hatley, M.E.; Galindo, R.L. Probing for a deeper understanding of rhabdomyosarcoma: Insights from complementary model systems. Nat. Rev. Cancer 2015, 15, 426–439. [Google Scholar] [CrossRef] [Green Version]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Primers 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Hinson, A.R.P.; Jones, R.; Lisa, L.E.; Belyea, B.C.; Barr, F.G.; Linardic, C.M. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: Utility and pitfalls. Front. Oncol. 2013, 3, 183. [Google Scholar] [CrossRef] [Green Version]
- Rossi, F.; Legnini, I.; Megiorni, F.; Colantoni, A.; Santini, T.; Morlando, M.; di Timoteo, G.; Dattilo, D.; Dominici, C.; Bozzoni, I. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene 2019, 38, 3843–3854. [Google Scholar] [CrossRef] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of M6A and M6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Bajno, L.; Peng, X.R.; Schreiber, A.D.; Moore, H.P.; Trimble, W.S.; Grinstein, S. Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J. Cell Biol. 2000, 149, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maass, P.G.; Glažar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef]
- Xu, N.; Lao, Y.; Zhang, Y.; Gillespie, D.A. Akt: A double-edged sword in cell proliferation and genome stability. J. Oncol. 2012, 2012, 951724. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, F.; Centrón-Broco, A.; Dattilo, D.; Di Timoteo, G.; Guarnacci, M.; Colantoni, A.; Beltran Nebot, M.; Bozzoni, I. CircVAMP3: A circRNA with a Role in Alveolar Rhabdomyosarcoma Cell Cycle Progression. Genes 2021, 12, 985. https://doi.org/10.3390/genes12070985
Rossi F, Centrón-Broco A, Dattilo D, Di Timoteo G, Guarnacci M, Colantoni A, Beltran Nebot M, Bozzoni I. CircVAMP3: A circRNA with a Role in Alveolar Rhabdomyosarcoma Cell Cycle Progression. Genes. 2021; 12(7):985. https://doi.org/10.3390/genes12070985
Chicago/Turabian StyleRossi, Francesca, Alvaro Centrón-Broco, Dario Dattilo, Gaia Di Timoteo, Marco Guarnacci, Alessio Colantoni, Manuel Beltran Nebot, and Irene Bozzoni. 2021. "CircVAMP3: A circRNA with a Role in Alveolar Rhabdomyosarcoma Cell Cycle Progression" Genes 12, no. 7: 985. https://doi.org/10.3390/genes12070985
APA StyleRossi, F., Centrón-Broco, A., Dattilo, D., Di Timoteo, G., Guarnacci, M., Colantoni, A., Beltran Nebot, M., & Bozzoni, I. (2021). CircVAMP3: A circRNA with a Role in Alveolar Rhabdomyosarcoma Cell Cycle Progression. Genes, 12(7), 985. https://doi.org/10.3390/genes12070985





