A Rapid and Cost-Effective Identification of Invertebrate Pests at the Borders Using MinION Sequencing of DNA Barcodes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. DNA Extraction
2.3. Amplification of DNA Barcode
2.4. Sequencing COI Amplicons
2.5. Bioinformatic Analysis
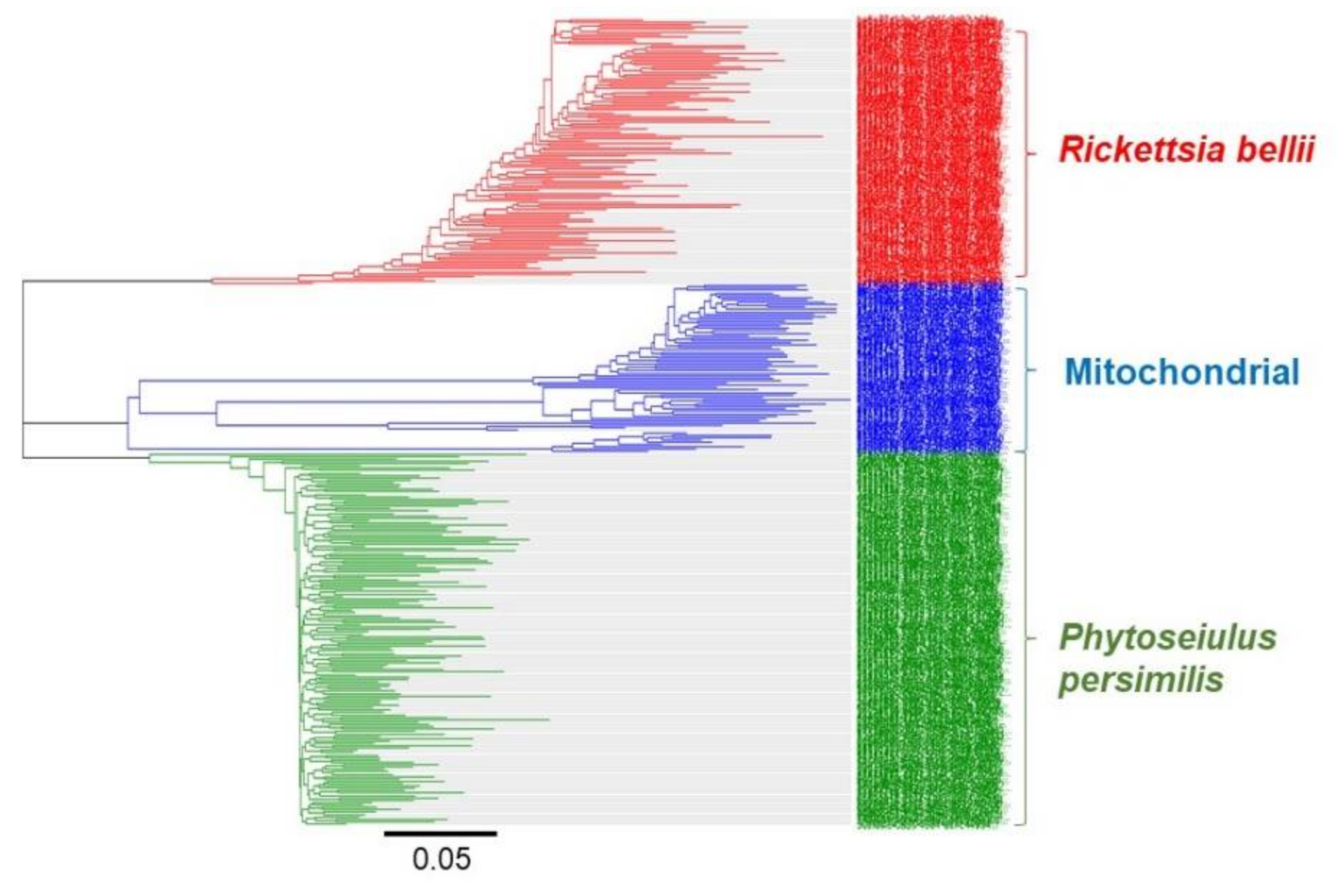
3. Results and Discussion
3.1. Identification of Invertebrates
3.2. MinION-Based Barcode Sequencing for Surveillance
3.3. Comparison of MinION Rapid Barcode Sequencing to Existing Sangar Sequencing and Morphological Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Dias-Lopes, C.; Paiva, A.L.; Guerra-Duarte, C.; Molina, F.; Felicori, L. Venomous arachnid diagnostic assays, lessons from past attempts. Toxins 2018, 10, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, D.A. The Current and Potential Costs of Invertebrate Pests in Grain Crops; Grain Research & Development Corporation: Canberra, Australia, 2013. [Google Scholar]
- Scheffers, B.R.; Joppa, L.N.; Pimm, S.L.; Laurance, W.F. What we know and don’t know about Earth’s missing biodiversity. Trends Ecol. Evol. 2012, 27, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Diaz, J.H. Mite-Transmitted Dermatoses and Infectious Diseases in Returning Travelers. J. Travel Med. 2010, 17, 21–31. [Google Scholar] [CrossRef]
- Pentinsaari, M.; Salmela, H.; Mutanen, M.; Roslin, T. Molecular evolution of a widely-adopted taxonomic marker (COI) across the animal tree of life. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Piper, A.M.; Batovska, J.; Cogan, N.O.I.; Weiss, J.; Cunningham, J.P.; Rodoni, B.C.; Blacket, M.J. Prospects and challenges of implementing DNA meta barcoding for high-throughput insect surveillance. GigaScience 2019, 8, giz092. [Google Scholar] [CrossRef]
- Franco-Sierra, N.D.; Díaz-Nieto, J.F. Rapid mitochondrial genome sequencing based on Oxford Nanopore Sequencing and a proxy for vertebrate species identification. Ecol. Evol. 2020, 10, 3544–3560. [Google Scholar] [CrossRef]
- Oppenheim, S.; Cao, X.; Rueppel, O.; Krongdang, S.; Phokasem, P.; DeSalle, R.; Goodwin, S.; Xing, J.; Chantawannakul, P.; Rosenfeld, J.A. Whole genome sequencing and assembly of the Asian honey bee Apis dorsata. Genome Biol. Evol. 2020, 12, 3677–3683. [Google Scholar] [CrossRef] [Green Version]
- Bronzato Badial, A.; Sherman, D.; Stone, A.; Gopakumar, A.; Wilson, V.; Schneider, W.; King, J. Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis. 2018, 102, 1648–1652. [Google Scholar] [CrossRef] [Green Version]
- Boykin, L.M.; Sseruwagi, P.; Alicai, T.; Ateka, E.; Mohammed, I.U.; Stanton, J.-A.L.; Kayuki, C.; Mark, D.; Fute, T.; Erasto, J. Tree Lab: Portable genomics for early detection of plant viruses and pests in Sub-Saharan Africa. Genes 2019, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Srivathsan, A.; Hartop, E.; Puniamoorthy, J.; Lee, W.T.; Kutty, S.N.; Kurina, O.; Meier, R. Rapid, large-scale species discovery in hyperdiverse taxa using 1D MinION sequencing. BMC Biol. 2019, 17, 96. [Google Scholar] [CrossRef] [Green Version]
- Srivathsan, A.; Baloğlu, B.; Wang, W.; Tan, W.X.; Bertrand, D.; Ng, A.H.; Boey, E.J.; Koh, J.J.; Nagarajan, N.; Meier, R. A Min ION™-based pipeline for fast and cost-effective DNA barcoding. Mol. Ecol. Resour. 2018, 18, 1035–1049. [Google Scholar] [CrossRef]
- Maestri, S.; Cosentino, E.; Paterno, M.; Freitag, H.; Garces, J.M.; Marcolungo, L.; Alfano, M.; Njunjić, I.; Schilthuizen, M.; Slik, F. A rapid and accurate MinION-based workflow for tracking species biodiversity in the field. Genes 2019, 10, 468. [Google Scholar] [CrossRef] [Green Version]
- Lienhard, A.; Schäffer, S. Extracting the invisible: Obtaining high quality DNA is a challenging task in small arthropods. PeerJ 2019, 7, e6753. [Google Scholar] [CrossRef] [PubMed]
- Oppert, B.; Stoss, S.; Monk, A.; Smith, T. Optimized Extraction of Insect Genomic DNA for Long-Read Sequencing. Methods Protoc. 2019, 2, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porco, D.; Rougerie, R.; Deharveng, L.; Hebert, P.J.M.E.R. Coupling non-destructive DNA extraction and voucher retrieval for small soft-bodied Arthropods in a high-throughput context: The example of Collembola. Mol. Ecol. Resour. 2010, 10, 942–945. [Google Scholar] [CrossRef]
- Ahaniazad, M.; Bagheri, M.; Roumi, V.; Al Akrami, M.; Protection, P. An efficient and non-destructive DNA extraction method for oribatid mites. Arch. Phytopathol. Plant Prot. 2018, 51, 187–196. [Google Scholar] [CrossRef]
- Hunter, S.J.; Goodall, T.I.; Walsh, K.A.; Owen, R.; Day, J.C. Nondestructive DNA extraction from blackflies (Diptera: Simuliidae): Retaining voucher specimens for DNA barcoding projects. Mol. Ecol. Resour. 2008, 8, 56–61. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit 1 from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kranzfelder, P.; Ekrem, T.; Stur, E. Trace DNA from insect skins: A comparison of five extraction protocols and direct PCR on chironomid pupal exuviae. Mol. Ecol. Resour. 2016, 16, 353–363. [Google Scholar] [CrossRef]
- Athanassiou, C.G.; Phillips, T.W.; Wakil, W. Biology and control of the khapra beetle, Trogoderma granarium, a major quarantine threat to global food security. Annu. Rev. Entomol. 2019, 64, 131–148. [Google Scholar] [CrossRef]
- Olson, R.L.; Farris, R.E.; Barr, N.B.; Cognato, A.I. Molecular identification of Trogoderma granarium (Coleoptera: Dermestidae) using the 16s gene. J. Pest Sci. 2014, 87, 701–710. [Google Scholar] [CrossRef]
- Ye, S.-S.; Zhang, Z.-Q.J.S.; Acarology, A. Age and size at maturity in Tyrophagus curvipenis (Acari: Acaridae) when fed on three different diets. Syst. Appl. Acarol. 2014, 19, 506–512. [Google Scholar]
- Murillo, P.; Klimov, P.; Hubert, J.; OConnor, B.J.E.; Acarology, A. Investigating species boundaries using DNA and morphology in the mite Tyrophagus curvipenis (Acari: Acaridae), an emerging invasive pest, with a molecular phylogeny of the genus Tyrophagus. Exp. Appl. Acarol. 2018, 75, 167–189. [Google Scholar] [CrossRef] [PubMed]
- Akyazi, R. First report of Aculops lycopersici (Tryon, 1917)(Acari: Eriophyidae) on Pepino in Turkey. J. Entomol. Acarol. Res. 2012, 44, e20. [Google Scholar] [CrossRef] [Green Version]
- Aysan, E.; Nabi, A.K.J.A. Tritrophic relationships among tomato cultivars, the rust mite, Aculops lycopersici (Massee)(Eriophyidae), and its predators. Acarologia 2018, 58, 5–17. [Google Scholar]
- Attia, S.; Grissa, K.L.; Lognay, G.; Bitume, E.; Hance, T.; Mailleux, A.C. A review of the major biological approaches to control the worldwide pest Tetranychus urticae (Acari: Tetranychidae) with special reference to natural pesticides. J. Pest Sci. 2013, 86, 361–386. [Google Scholar] [CrossRef]
- Ristyadi, D.; He, X.Z.; Wang, Q.J.S.; Acarology, A. Dynamics of life history traits in Tetranychus ludeni Zacher in response to fluctuating temperatures. Syst. Appl. Acarol. 2019, 24, 2272–2277. [Google Scholar]
- Adango, E.; Onzo, A.; Hanna, R.; Atachi, P.; James, B. Comparative demography of the spider mite, Tetranychus ludeni, on two host plants in West Africa. J. Insect Sci. 2006, 6, 49. [Google Scholar] [CrossRef] [Green Version]
- Seeman, O.D.; Beard, J.J. Identification of exotic pest and Australian native and naturalised species of Tetranychus (Acari: Tetranychidae). Zootaxa 2011, 2961, 1–72. [Google Scholar] [CrossRef] [Green Version]
- Fathipour, Y.; Karimi, M.; Farazmand, A.; Talebi, A.A. Age-specific functional response and predation capacity of Phytoseiulus persimilis (Phytoseiidae) on the two-spotted spider mite. Acarologia 2017, 58, 31–40. [Google Scholar]
- Derocles, S.A.; Le Ralec, A.; Plantegenest, M.; Chaubet, B.; Cruaud, C.; Cruaud, A.; Rasplus, J.Y. Identification of molecular markers for DNA barcoding in the Aphidiinae (Hym. Braconidae). Mol. Ecol. Resour. 2012, 12, 197–208. [Google Scholar] [CrossRef]
- EPPO. PM 7/129 (1) DNA barcoding as an identification tool for a number of regulated pests. EPPO Bull. 2016, 46, 501–537. [Google Scholar] [CrossRef] [Green Version]
- Seah, A.; Lim, M.C.; McAloose, D.; Prost, S.; Seimon, T.A. MinION-based DNA barcoding of preserved and non-invasively collected wildlife samples. Genes 2020, 11, 445. [Google Scholar] [CrossRef] [Green Version]
- Madden, M.J.; Young, R.G.; Brown, J.W.; Miller, S.E.; Frewin, A.J.; Hanner, R.H. Using DNA barcoding to improve invasive pest identification at US ports-of-entry. PLoS ONE 2019, 14, e0222291. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence |
|---|---|
| LCO1490 (Forward) | 5′-GGTCAACAAATCATAAAGATATTGG-3′ |
| HC02198 (Reverse) | 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ |
| Number | Developmental Stage | Initial Determination by Microscopic Methods | Determination by MinION Barcode Sequencing | Comments |
|---|---|---|---|---|
| 1 | Adult | Trogoderma | Trogoderma variabile | Improved BDM* |
| 98.48% Id* | ||||
| 2 | Adult | Trogoderma | Trogoderma anthrenoides | Improved BDM* |
| 97.89% Id* | ||||
| 3 | Adult | Beetle | Trogoderma granarium | Improved BDM* |
| 97.92% Id* | ||||
| 4 | Adult | Trogoderma granarium | Trogoderma granarium | Supported BDM* |
| 98.37% Id* | ||||
| 5 | Adult | Franklinella occidentalis | Franklinella occidentalis | Supported BDM* |
| 98.05% Id* | ||||
| 6 | Larva | Thripidae | Franklinella occidentalis | Improved BDM* |
| 98.39% Id* | ||||
| 7 | Adult | Franklinella shultzei | Franklinella shultzei | Supported BDM* |
| 98.05% Id* | ||||
| 8 | Larva | Thripidae | Franklinella occidentalis | Improved BDM* |
| 99.55% Id* | ||||
| 9 | Larva | Thripidae | Franklinella occidentalis | Improved BDM* |
| 98.35% Id* | ||||
| 10 | Adult | Franklinella occidentalis | Franklinella occidentalis | Supported BDM* |
| 99.55% Id* | ||||
| 11 | Adult | Franklinella occidentalis | Franklinella occidentalis | Supported BDM* |
| 99.10% Id* | ||||
| 12 | Adult | Franklinella occidentalis | Franklinella occidentalis | Supported BDM* |
| 99.70% Id* | ||||
| 13 | Adult | Franklinella shultzei | Franklinella shultzei | Supported BDM* |
| 98.63% Id* | ||||
| 14 | Adult | Franklinella shultzei | Franklinella shultzei | Supported BDM* |
| 98.25% Id* | ||||
| 15 | Adult | Franklinella occidentalis | Franklinella occidentalis | Supported BDM* |
| 98.86% Id* | ||||
| 16 | Adult | Franklinella occidentalis | Franklinella occidentalis | Supported BDM* |
| 97.68% Id* |
| Number | Developmental Stage | Initial Determination by Microscopic Methods | Determination by MinION Sequencing of Barcode | Comments |
|---|---|---|---|---|
| 1 | Adult | Tyrophagus curvipenis | Tyrophagus curvipenis | Supported BDM* |
| 97.29% Id* | ||||
| 2 | Adult | Tetranychus urticae | Tetranychus urticae | Supported BDM* |
| 99.25% Id* | ||||
| 3 | Adult | Tetranychus ludeni | Tetranychus ludeni | Supported BDM* |
| 98.96% Id* | ||||
| 4 | Adult | Tetranychus evansi | Aculops lycopersici | Improved BDM* |
| 98.67% Id* | ||||
| 5 | Adult | Acari (Sub-class) | Phytoseiulus persimilis | Improved BDM* |
| 97.47% Id* | ||||
| 6 | Egg | Arachnida (Class) | Phytoseiulus persimilis | Improved BDM* |
| 97.40% Id* | ||||
| 7 | Egg | Not detected | Haplothrips sp | Improved BDM* |
| 98.92% Id* | ||||
| 8 | Egg | Insecta (Class) | Phloeonomus punctipennis | Improved BDM* |
| 97.19% Id* | ||||
| 9 | Egg | Not detected | Signiphora flavella | Improved BDM* |
| 98.93% Id* | ||||
| 10 | Egg | Blattodea (Order) | Choristima sp | Improved BDM* |
| 99.24% Id* |
| Number | Determination by Sanger Sequencing of Barcode | Determination by MinION Sequencing of Barcode | Database |
|---|---|---|---|
| 1 | Bactrocera xanthodes 99.88% Id* | Bactrocera xanthodes 99.24% Id* | NCBI |
| 2 | Aleuroctarthrus destructor 99.34% Id* | Aleuroctarthrus destructor 99.38% Id* | BOLD |
| 3 | Lepidoptera 100% Id* | Lepidoptera 100% Id* | BOLD |
| 4 | Spodoptera litura 100% Id* | Spodoptera litura 100% Id* | BOLD |
| 5 | Liriomyza trifolii 100% Id* | Liriomyza trifolii 99.64% Id* | BOLD |
| 6 | Helicoverpa armigera 99.85% Id* | Helicoverpa armigera 98.93% Id* | NCBI |
| 7 | Aleyrodidae 78.64% Id* | Aleyrodidae 77.4% Id* | BOLD |
| 8 | Helicoverpa armigera 100% Id* | Helicoverpa armigera 100% Id* | BOLD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abeynayake, S.W.; Fiorito, S.; Dinsdale, A.; Whattam, M.; Crowe, B.; Sparks, K.; Campbell, P.R.; Gambley, C. A Rapid and Cost-Effective Identification of Invertebrate Pests at the Borders Using MinION Sequencing of DNA Barcodes. Genes 2021, 12, 1138. https://doi.org/10.3390/genes12081138
Abeynayake SW, Fiorito S, Dinsdale A, Whattam M, Crowe B, Sparks K, Campbell PR, Gambley C. A Rapid and Cost-Effective Identification of Invertebrate Pests at the Borders Using MinION Sequencing of DNA Barcodes. Genes. 2021; 12(8):1138. https://doi.org/10.3390/genes12081138
Chicago/Turabian StyleAbeynayake, Shamila Weerakoon, Sonia Fiorito, Adrian Dinsdale, Mark Whattam, Bill Crowe, Kate Sparks, Paul Richard Campbell, and Cherie Gambley. 2021. "A Rapid and Cost-Effective Identification of Invertebrate Pests at the Borders Using MinION Sequencing of DNA Barcodes" Genes 12, no. 8: 1138. https://doi.org/10.3390/genes12081138
APA StyleAbeynayake, S. W., Fiorito, S., Dinsdale, A., Whattam, M., Crowe, B., Sparks, K., Campbell, P. R., & Gambley, C. (2021). A Rapid and Cost-Effective Identification of Invertebrate Pests at the Borders Using MinION Sequencing of DNA Barcodes. Genes, 12(8), 1138. https://doi.org/10.3390/genes12081138








