Glutamine Metabolism Regulators Associated with Cancer Development and the Tumor Microenvironment: A Pan-Cancer Multi-Omics Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Genome-Wide Analysis
2.3. Survival Analysis
2.4. Analysis of Immune Cell Infiltration
2.5. Drug Sensitivity Analysis
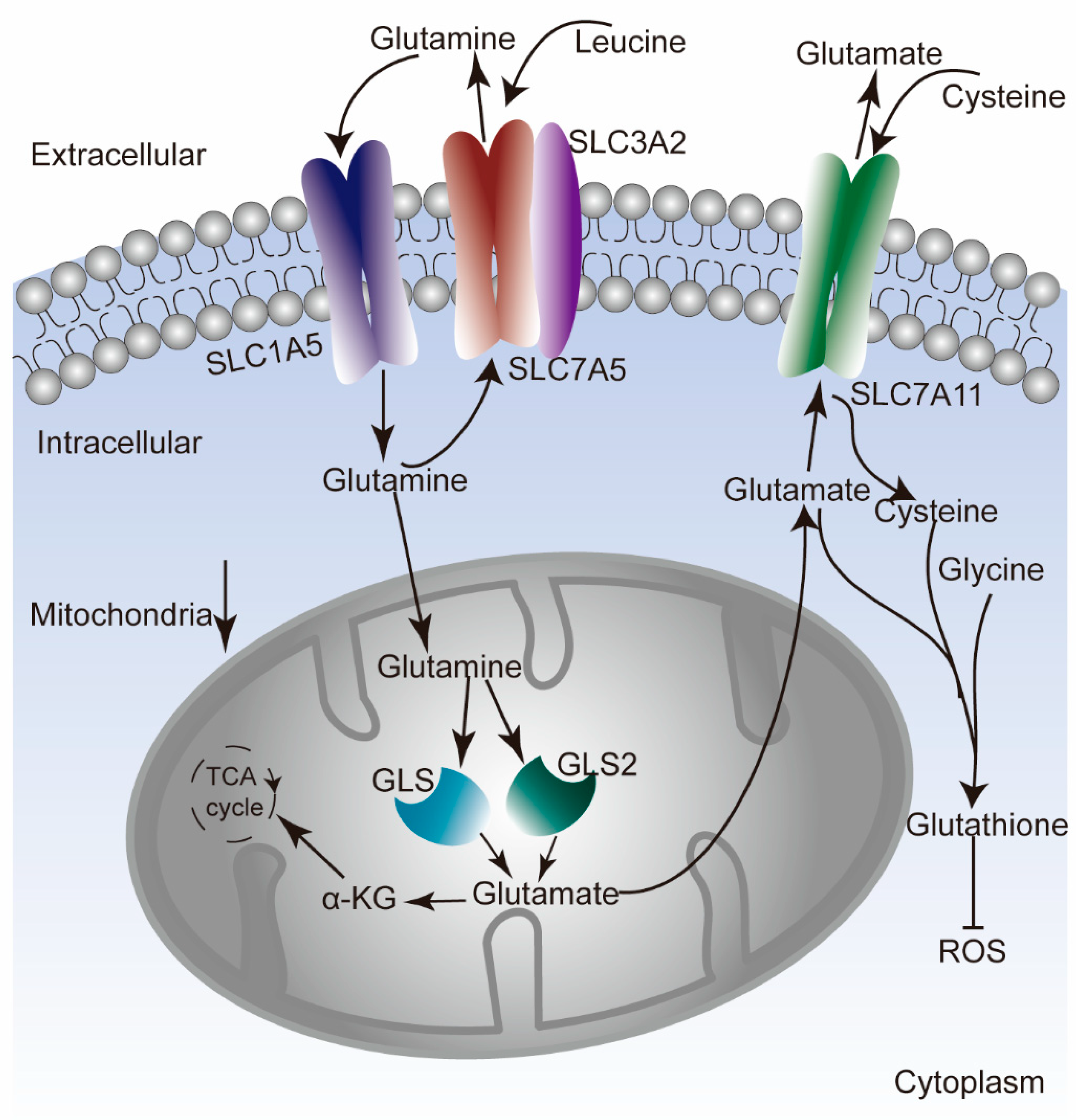
2.6. Analysis of Molecular Mechanisms
2.7. Statistical Analysis
3. Results
3.1. Aberrant Expression of GLNM Regulators in a Variety of Cancers
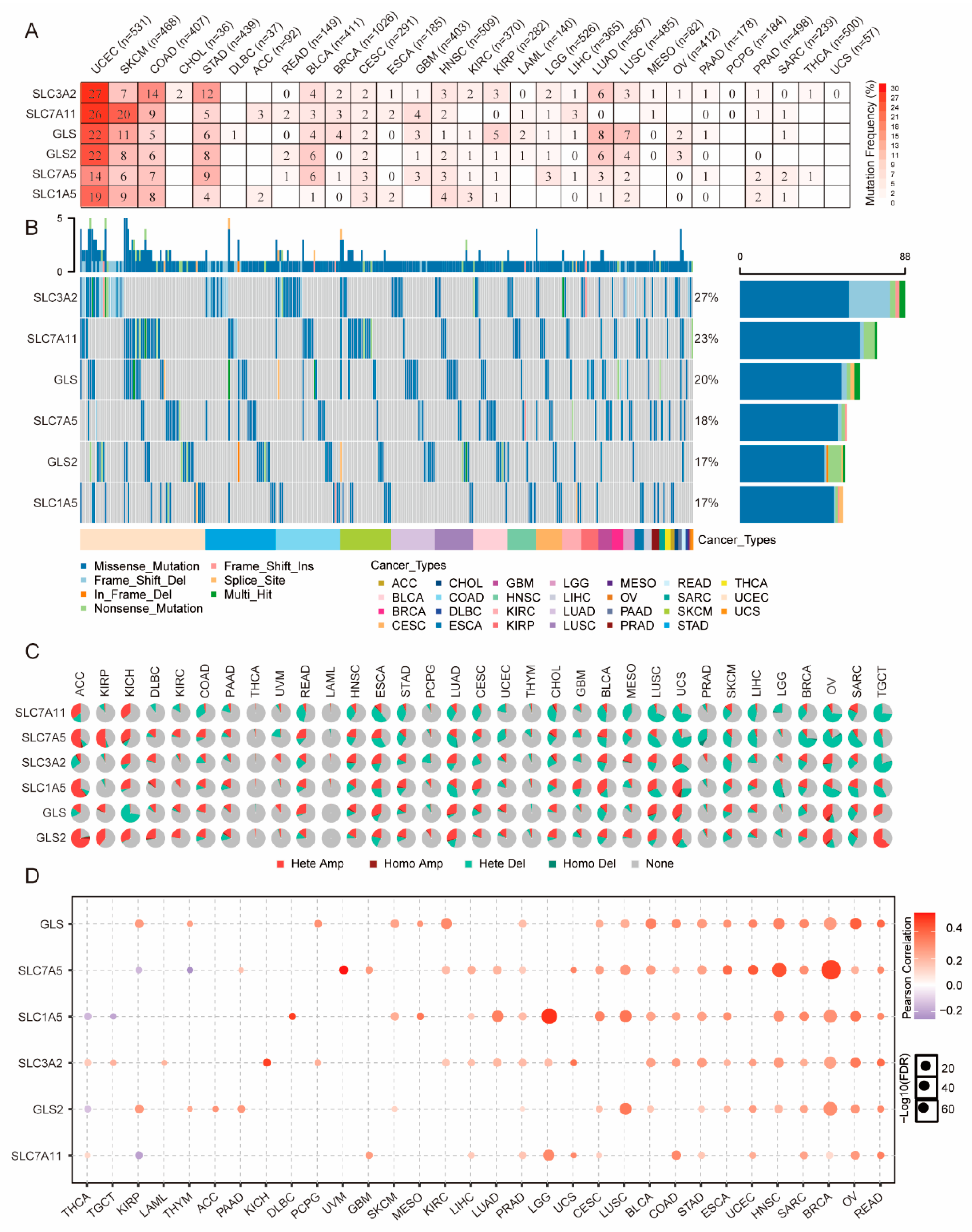
3.2. Genetic Alterations of GLNM Regulators in Various Cancers
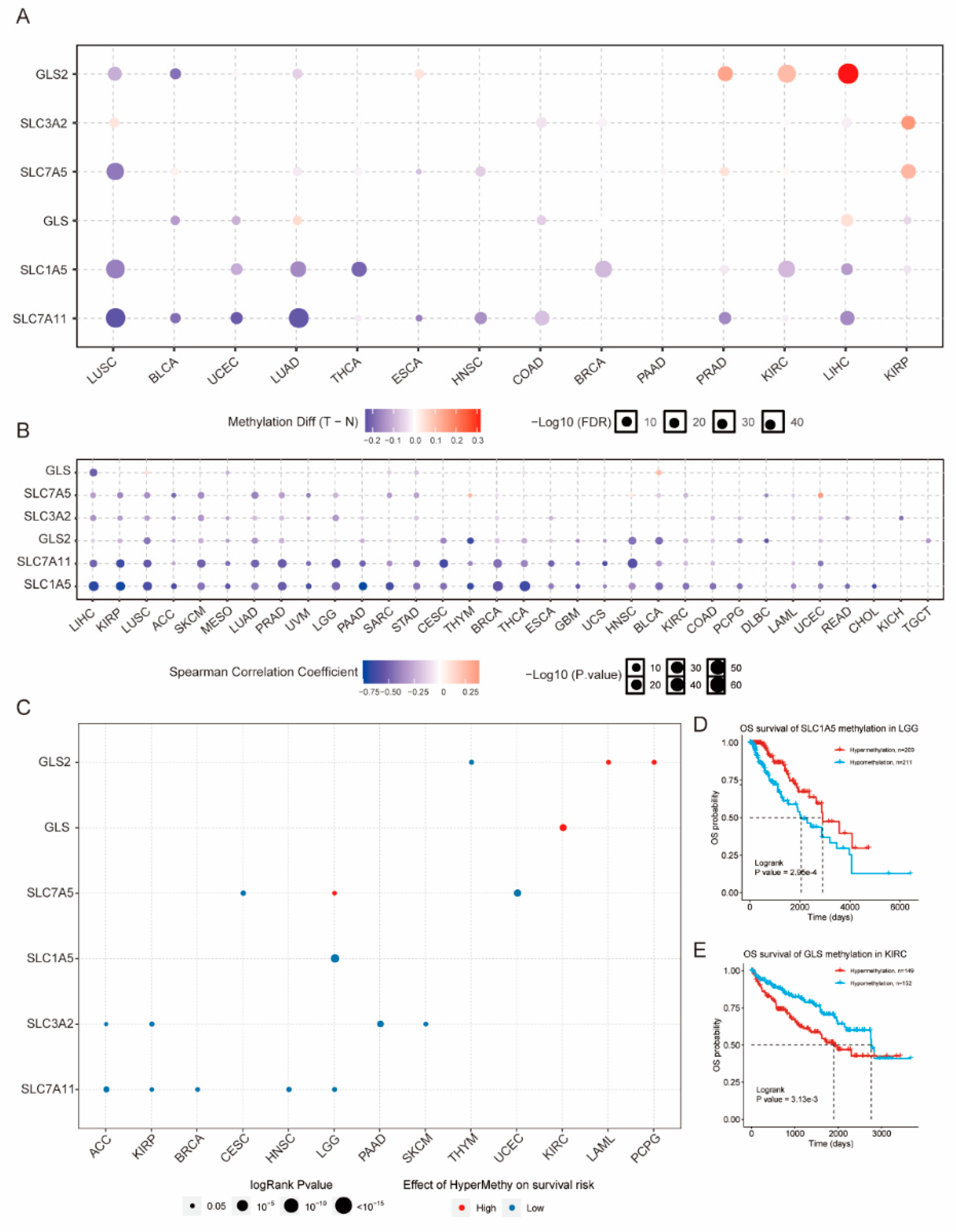
3.3. Epigenetic Alteration of GLNM Regulators in Various Cancers
3.4. Significant Correlation of GLNM Regulators with Survival
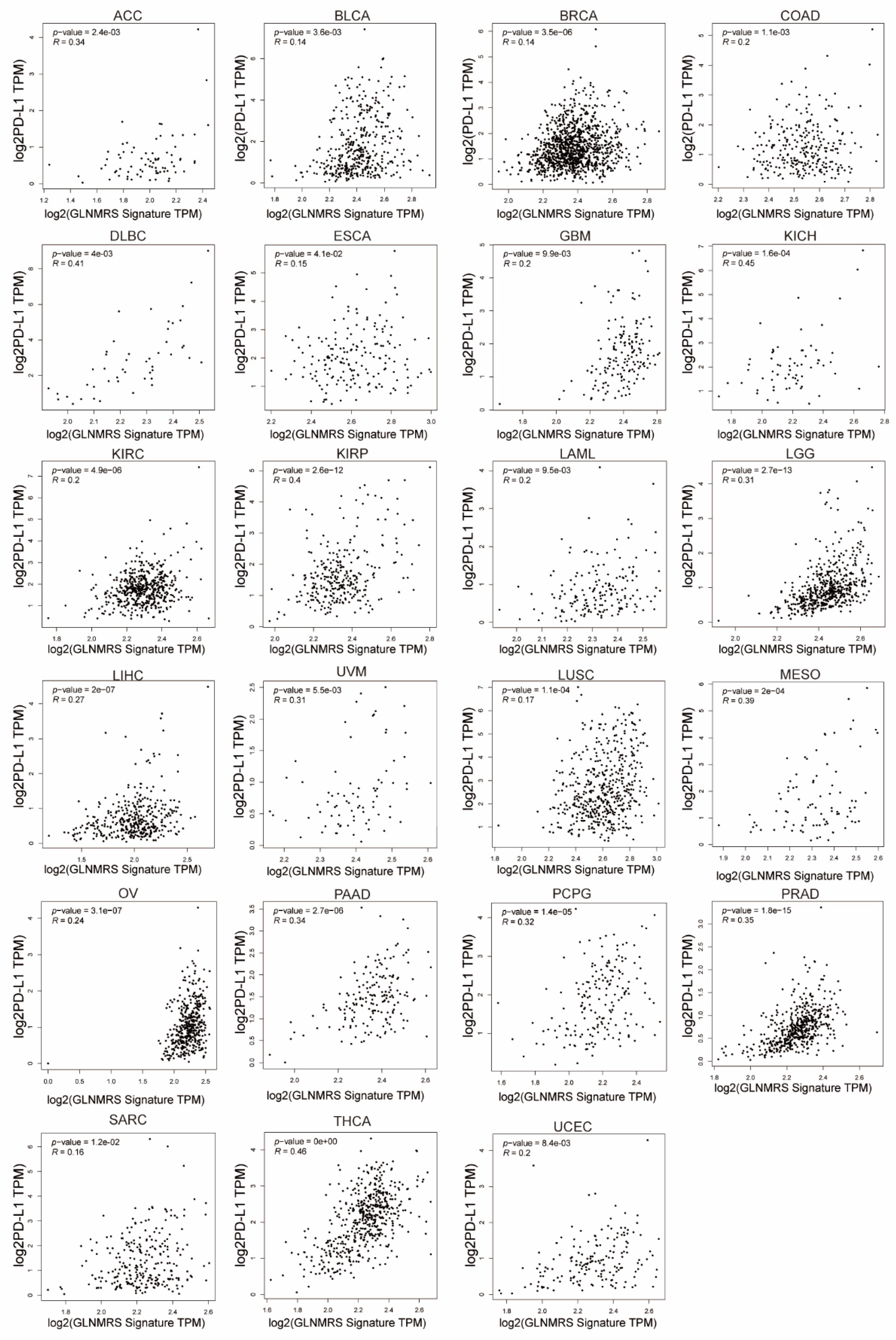
3.5. Association of Immune Cell Infiltration and PD-L1 with GLNM Regulators
3.6. Effects of Aberrant GLNM Regulator Expression on Drug Sensitivity
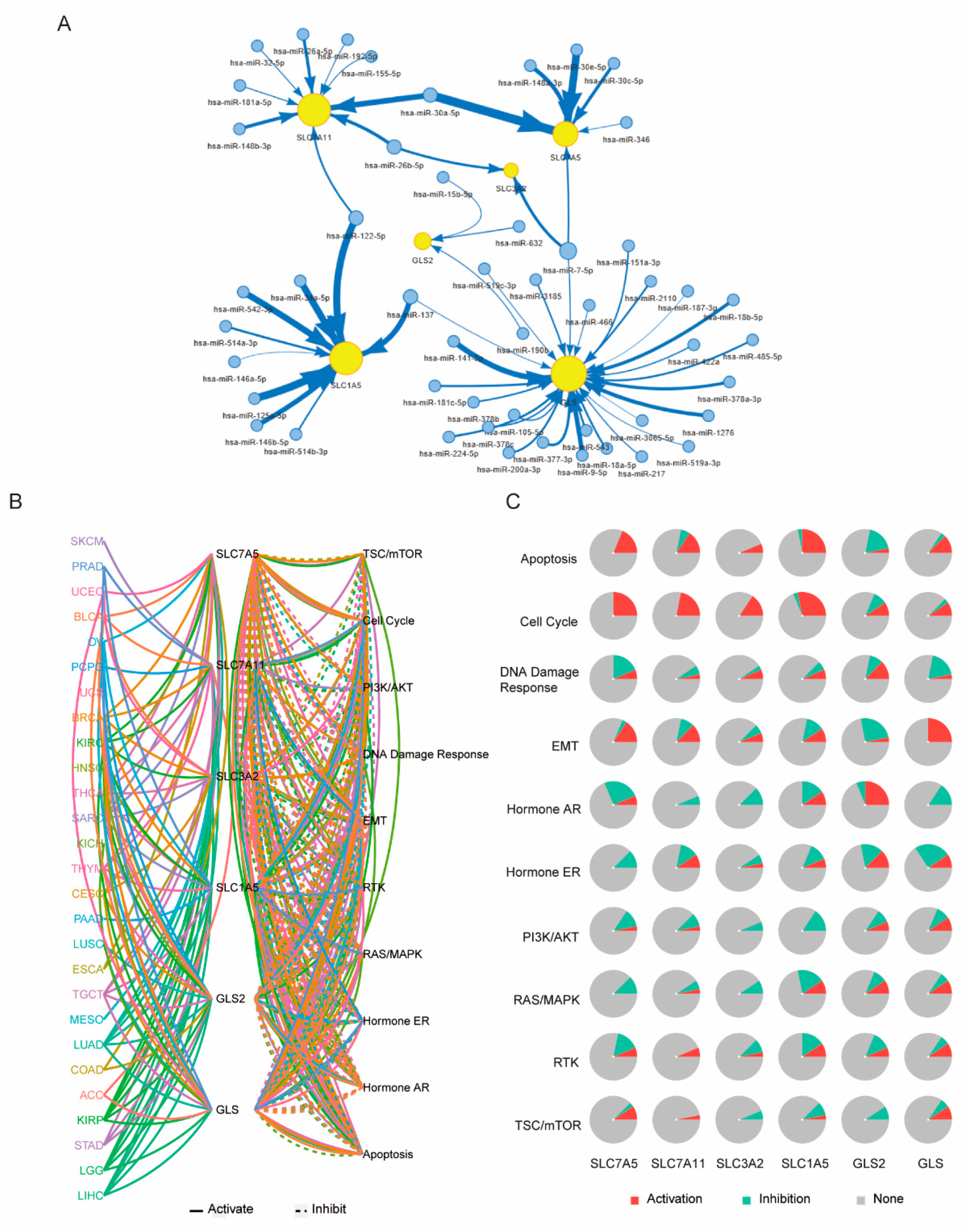
3.7. Underlying Molecular Mechanism of GLNM Regulator Alteration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, Y.E.M.; Alves, M.J.; Uno, M.; Moretti, I.F.; Trombetta-Lima, M.; de Siqueira Santos, S.; Dos Santos, A.F.; Arini, G.S.; Baptista, M.S.; Lerario, A.M.; et al. Glutaminolysis dynamics during astrocytoma progression correlates with tumor aggressiveness. Cancer Metab. 2021, 9, 18. [Google Scholar] [CrossRef]
- Bott, A.; Peng, I.-C.; Fan, Y.; Faubert, B.; Zhao, L.; Li, J.; Neidler, S.; Sun, Y.; Jaber, N.; Krokowski, D.; et al. Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation. Cell Metab. 2015, 22, 1068–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Still, E.R.; Yuneva, M. Hopefully devoted to Q: Targeting glutamine addiction in cancer. Br. J. Cancer 2017, 116, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Sayed, N.; Ditsworth, D.; Thompson, C.B. Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008, 18, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Bhutia, Y.D.; Babu, E.; Ramachandran, S.; Ganapathy, V. Amino Acid transporters in cancer and their relevance to “glutamine addiction”: Novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015, 75, 1782–1788. [Google Scholar] [CrossRef] [Green Version]
- Yanagida, O.; Kanai, Y.; Chairoungdua, A.; Kim, D.K.; Segawa, H.; Nii, T.; Cha, S.H.; Matsuo, H.; Fukushima, J.-I.; Fukasawa, Y.; et al. Human L-type amino acid transporter 1 (LAT1): Characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta (BBA) Biomembr. 2001, 1514, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Timmerman, L.A.; Holton, T.; Yuneva, M.; Louie, R.J.; Padró, M.; Daemen, A.; Hu, M.; Chan, D.A.; Ethier, S.P.; van’t Veer, L.J.; et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 2013, 24, 450–465. [Google Scholar] [CrossRef] [Green Version]
- Lv, H.; Zhen, C.; Liu, J.; Yang, P.; Hu, L.; Shang, P. Unraveling the Potential Role of Glutathione in Multiple Forms of Cell Death in Cancer Therapy. Oxidative Med. Cell. Longev. 2019, 2019, 3150145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Ding, Y.; Wang, X.; Lu, S.; Wang, C.; He, C.; Wang, L.; Piao, M.; Chi, G.; Luo, Y.; et al. Pseudolaric acid B triggers ferroptosis in glioma cells via activation of Nox4 and inhibition of xCT. Cancer Lett. 2018, 428, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Katt, W.; Lukey, M.; Cerione, R. A tale of two glutaminases: Homologous enzymes with distinct roles in tumorigenesis. Future Med. Chem. 2017, 9, 223–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Peña, E.; Arnold, J.; Shivakumar, V.; Joseph, R.; Vijay, G.V.; den Hollander, P.; Bhangre, N.; Allegakoen, P.; Prasad, R.; Conley, Z.; et al. The Epithelial to Mesenchymal Transition Promotes Glutamine Independence by Suppressing GLS2 Expression. Cancers 2019, 11, 1610. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Dai, Z.; Barbacioru, C.; Sadée, W. Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res. 2005, 65, 7446–7454. [Google Scholar] [CrossRef] [Green Version]
- Lo, M.; Ling, V.; Wang, Y.; Gout, P.W. The xc-cystine/glutamate antiporter: A mediator of pancreatic cancer growth with a role in drug resistance. Br. J. Cancer 2008, 99, 464–472. [Google Scholar] [CrossRef] [Green Version]
- Drayton, R.M.; Dudziec, E.; Peter, S.; Bertz, S.; Hartmann, A.; Bryant, H.E.; Catto, J.W. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin. Cancer Res. 2014, 20, 1990–2000. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-J.; Hu, F.-F.; Xia, M.-X.; Han, L.; Zhang, Q.; Guo, A.-Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Andersen, P.K.; Gill, R.D. Cox’s Regression Model for Counting Processes: A Large Sample Study. Ann. Stat. 1982, 10, 1100–1120. [Google Scholar] [CrossRef]
- Harrington, D.P.; Fleming, T.R. A class of rank test procedures for censored survival data. Biometrika 1982, 69, 553–566. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.Y.; Wang, H.; Guo, A.Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. 2020, 7, 1902880. [Google Scholar] [CrossRef] [Green Version]
- Akbani, R.; Ng, P.K.S.; Werner, H.M.J.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.-Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef]
- Ye, Y.; Xiang, Y.; Ozguc, F.M.; Kim, Y.; Liu, C.-J.; Park, P.K.; Hu, Q.; Diao, L.; Lou, Y.; Lin, C.; et al. The Genomic Landscape and Pharmacogenomic Interactions of Clock Genes in Cancer Chronotherapy. Cell Syst. 2018, 6, 314–328.e312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaira, K.; Oriuchi, N.; Imai, H.; Shimizu, K.; Yanagitani, N.; Sunaga, N.; Hisada, T.; Tanaka, S.; Ishizuka, T.; Kanai, Y.; et al. l-type amino acid transporter 1 and CD98 expression in primary and metastatic sites of human neoplasms. Cancer Sci. 2008, 99, 2380–2386. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Lane, A.N.; Robertson, B.; Kemp, S.R.M.; Liu, Y.; Hill, B.; Dean, D.C.; Clem, B.F. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 2014, 33, 556–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewerenz, J.; Hewett, S.J.; Huang, Y.; Lambros, M.; Gout, P.W.; Kalivas, P.W.; Massie, A.; Smolders, I.; Methner, A.; Pergande, M. The cystine/glutamate antiporter system x(c)(-) in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 2013, 18, 522–555. [Google Scholar] [CrossRef] [Green Version]
- Deuschle, F.-C.; Morath, V.; Schiefner, A.; Brandt, C.; Ballke, S.; Reder, S.; Steiger, K.; Schwaiger, M.; Weber, W.; Skerra, A. Development of a high affinity Anticalin® directed against human CD98hc for theranostic applications. Theranostics 2020, 10, 2172–2187. [Google Scholar] [CrossRef]
- Matés, J.M.; Campos-Sandoval, J.A.; Márquez, J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 158–164. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wang, C.; Chen, M.; Cao, J.; Zhong, Y.; Chen, L.; Shen, H.-M.; Xia, D. Epigenetic silencing of glutaminase 2 in human liver and colon cancers. BMC Cancer 2013, 13, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengupta, D.; Cassel, T.; Teng, K.-Y.; Aljuhani, M.; Chowdhary, V.K.; Hu, P.; Zhang, X.; Fan, T.W.-M.; Ghoshal, K. Regulation of hepatic glutamine metabolism by miR-122. Mol. Metab. 2020, 34, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Tanaka, T.; Poyurovsky, M.V.; Nagano, H.; Mayama, T.; Ohkubo, S.; Lokshin, M.; Hosokawa, H.; Nakayama, T.; Suzuki, Y.; et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. USA 2010, 107, 7461–7466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, T.-C.; Chen, C.-K.; Hua, K.-T.; Yu, P.; Lee, W.-J.; Chen, M.-W.; Jeng, Y.-M.; Chien, M.-H.; Kuo, K.-T.; Hsiao, M.; et al. Glutaminase 2 stabilizes Dicer to repress Snail and metastasis in hepatocellular carcinoma cells. Cancer Lett. 2016, 383, 282–294. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhao, Y.; Yue, X.; Zhu, Y.; Wang, X.; Wu, H.; Blanco, F.; Li, S.; Bhanot, G.; et al. Author response: Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. eLife 2015, 5, e10727. [Google Scholar] [CrossRef] [PubMed]
- Lukey, M.; Cluntun, A.; Katt, W.P.; Lin, M.-C.J.; Druso, J.E.; Ramachandran, S.; Erickson, J.W.; Le, H.; Wang, Z.-E.; Blank, B.; et al. Liver-Type Glutaminase GLS2 Is a Druggable Metabolic Node in Luminal-Subtype Breast Cancer. Cell Rep. 2019, 29, 76–88.e77. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.-W.; Wu, W.-C.; Chen, H.-T.; Xu, Y.-T.; Yang, Y.-Y.; Chen, J.; Yu, X.-J.; Wang, Z.; Shuang, Z.-Y.; Zheng, L. Glutamine Deprivation Promotes the Generation and Mobilization of MDSCs by Enhancing Expression of G-CSF and GM-CSF. Front. Immunol. 2021, 11, 616367. [Google Scholar] [CrossRef] [PubMed]
- El Ansari, R.; Craze, M.L.; Althobiti, M.; Alfarsi, L.; Ellis, I.O.; Rakha, E.A.; Green, A.R. Enhanced glutamine uptake influences composition of immune cell infiltrates in breast cancer. Br. J. Cancer 2020, 122, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Li, C.; Zhang, Z.; Liang, Y.; Liang, Z.; Chen, Y.; Wang, L.; Li, D.; Zeng, M.; Shan, W.; et al. Targeted Glucose or Glutamine Metabolic Therapy Combined With PD-1/PD-L1 Checkpoint Blockade Immunotherapy for the Treatment of Tumors—Mechanisms and Strategies. Front. Oncol. 2021, 11, 697894. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.-K.; Park, M.; Lee, S.; Yun, J.W.; Lee, J.; Kim, J.S.; Cho, S.J.; Jeon, H.-J.; Lee, I.-K.; Choi, Y.-K.; et al. Inhibition of Glutamine Utilization Synergizes with Immune Checkpoint Inhibitor to Promote Antitumor Immunity. Mol. Cell 2020, 80, 592–606.e598. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Liu, M.; Huang, W.; Chen, X.; Zhang, B.; Zhang, T.; Wang, Y.; Liu, D.; Xie, M.; Ji, X.; et al. IL-1β-induced Elevation of SLC7A11 Promotes Hepatocellular Carcinoma Metastasis through Upregulating PD-L1 and CSF1. Hepatology 2021. epub ahead of print. [Google Scholar] [CrossRef]
- Pham, A.N.; Blower, P.E.; Alvarado, O.; Ravula, R.; Gout, P.W.; Huang, Y. Pharmacogenomic approach reveals a role for the x(c)-cystine/glutamate antiporter in growth and celastrol resistance of glioma cell lines. J. Pharmacol. Exp. Ther. 2010, 332, 949–958. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, K.; Watanabe, M.; Boku, S.; Sukeno, M.; Morita, M.; Kondo, H.; Sakaguchi, K.; Taguchi, T.; Sakai, A.T. xCT Inhibition Increases Sensitivity to Vorinostat in a ROS-Dependent Manner. Cancers 2020, 12, 827. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Sasayama, T.; Irino, Y.; Takata, K.; Nagashima, H.; Satoh, N.; Kyotani, K.; Mizowaki, T.; Imahori, T.; Ejima, Y.; et al. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J. Clin. Investig. 2015, 125, 1591–1602. [Google Scholar] [CrossRef]
- Momcilovic, M.; Bailey, S.T.; Lee, J.T.; Fishbein, M.C.; Braas, D.; Go, J.; Graeber, T.G.; Parlati, F.; Demo, S.; Li, R.; et al. The GSK3 Signaling Axis Regulates Adaptive Glutamine Metabolism in Lung Squamous Cell Carcinoma. Cancer Cell 2018, 33, 905–921.e905. [Google Scholar] [CrossRef] [Green Version]
- Boku, S.; Watanabe, M.; Sukeno, M.; Yaoi, T.; Hirota, K.; Iizuka-Ohashi, M.; Itoh, K.; Sakai, T. Deactivation of Glutaminolysis Sensitizes PIK3CA-Mutated Colorectal Cancer Cells to Aspirin-Induced Growth Inhibition. Cancers 2020, 12, 1097. [Google Scholar] [CrossRef]
- Li, Y.; Umbach, D.M.; Krahn, J.M.; Shats, I.; Li, X.; Li, L. Predicting tumor response to drugs based on gene-expression biomarkers of sensitivity learned from cancer cell lines. BMC Genom. 2021, 22, 272. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Du, K.; Li, S.; Lu, L.; Mei, J.; Lin, W.; Deng, M.; Wei, W.; Guo, R. Glutamine Metabolism Regulators Associated with Cancer Development and the Tumor Microenvironment: A Pan-Cancer Multi-Omics Analysis. Genes 2021, 12, 1305. https://doi.org/10.3390/genes12091305
Zou J, Du K, Li S, Lu L, Mei J, Lin W, Deng M, Wei W, Guo R. Glutamine Metabolism Regulators Associated with Cancer Development and the Tumor Microenvironment: A Pan-Cancer Multi-Omics Analysis. Genes. 2021; 12(9):1305. https://doi.org/10.3390/genes12091305
Chicago/Turabian StyleZou, Jingwen, Kunpeng Du, Shaohua Li, Lianghe Lu, Jie Mei, Wenping Lin, Min Deng, Wei Wei, and Rongping Guo. 2021. "Glutamine Metabolism Regulators Associated with Cancer Development and the Tumor Microenvironment: A Pan-Cancer Multi-Omics Analysis" Genes 12, no. 9: 1305. https://doi.org/10.3390/genes12091305





