Microarray Analysis Revealed Inflammatory Transcriptomic Changes after LSL60101 Treatment in 5XFAD Mice Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Brain Processing and RNA Extraction
2.3. Real-time Quantitative PCR Array
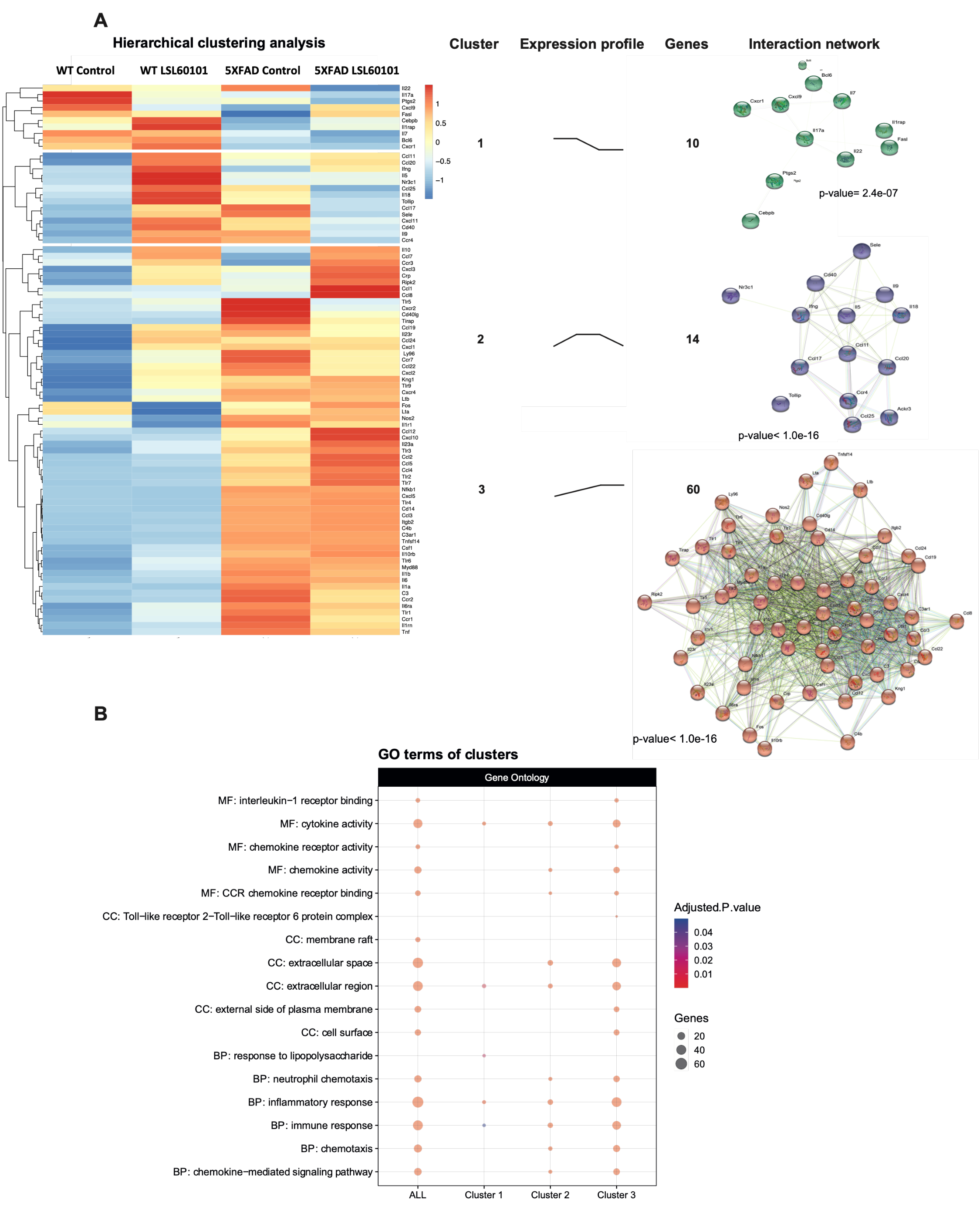
2.4. Hierarchical Clustering
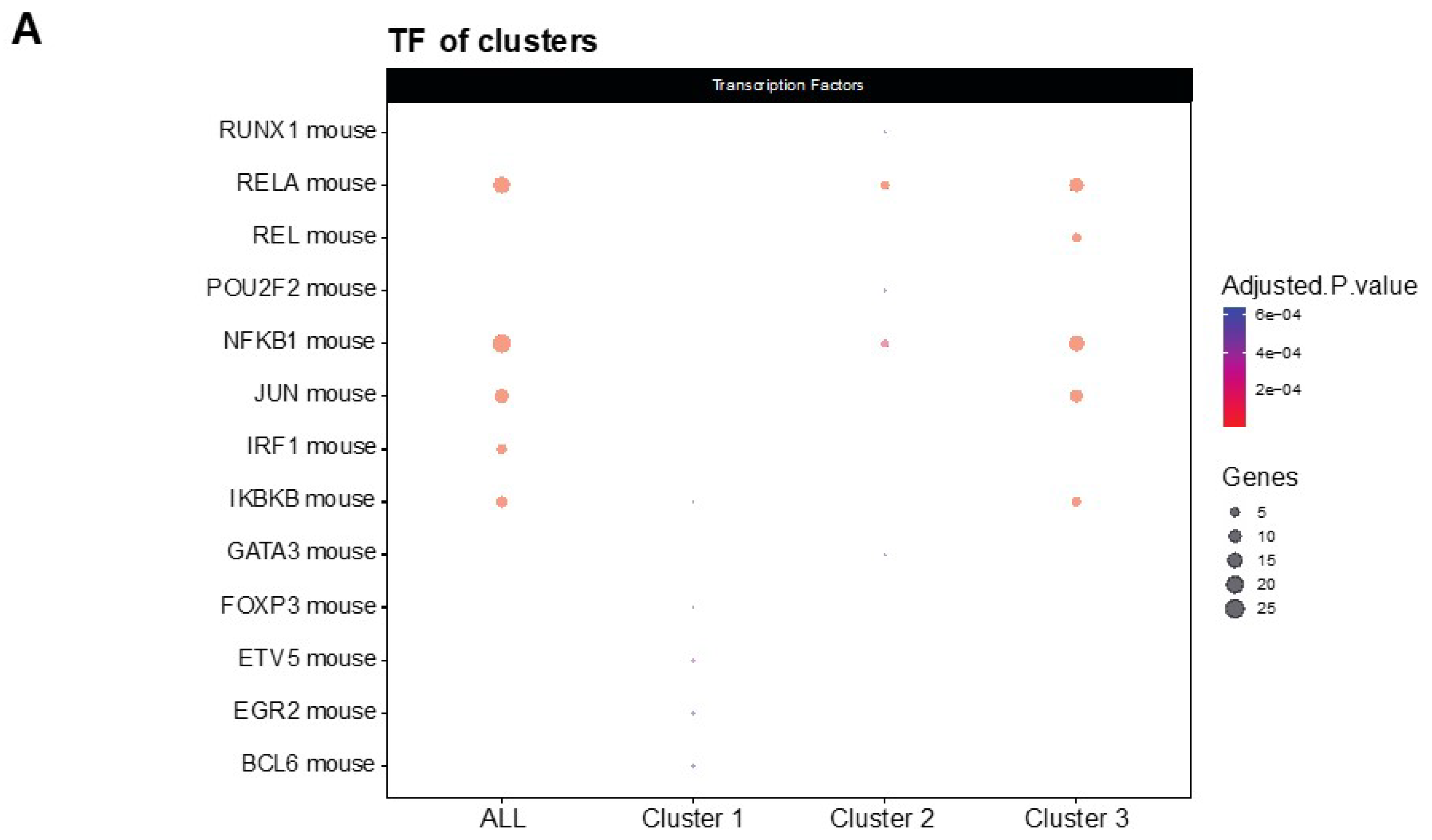
2.5. Protein–Protein Interaction Network and Functional Annotation
2.6. Gene Expression Validation with Real-Time Quantitative PCR
2.7. Statistical Analysis
3. Results
3.1. LSL60101 Treatment Regulates Genes Associated with the Inflammatory Response
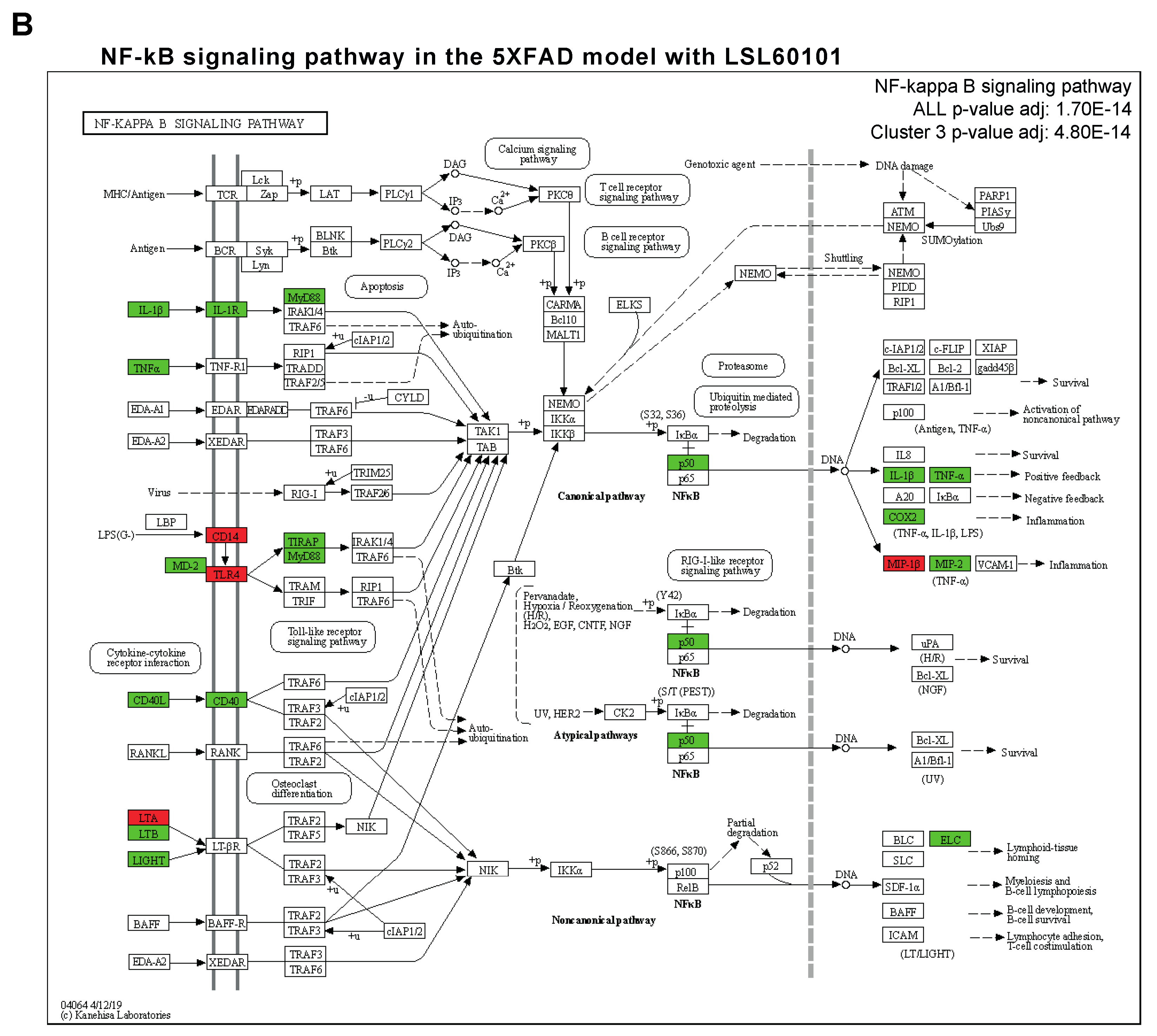
3.2. NF-κβ Pathway Regulates the Inflammatory Response in LSL60101 Treatment
3.3. Validation of a Representative Subset of Genes Involved in Neuroinflammation and AD
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bousquet, P.; Feldman, J.; Schwartz, J. Central Cardiovascular Effects of Alpha Adrenergic Drugs: Differences between Catecholamines and Imidazolines. J. Pharmacol. Exp. Ther. 1984, 230, 232–236. [Google Scholar]
- Ernsberger, P.; Meeley, M.P.; Mann, J.J.; Reis, D.J. Clonidine Binds to Imidazole Binding Sites as Well as Alpha 2-Adrenoceptors in the Ventrolateral Medulla. Eur. J. Pharmacol. 1987, 134, 1–13. [Google Scholar] [CrossRef]
- Diamant, S.; Eldar-Geva, T.; Atlas, D. Imidazoline Binding Sites in Human Placenta: Evidence for Heterogeneity and a Search for Physiological Function. Br. J. Pharmacol. 1992, 106, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regunathan, S.; Reis, D.J. Imidazoline Receptors and Their Endogenous Ligands. Annu. Rev. Pharmacol. Toxicol. 1996, 36, 511–544. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.L.; Brown, C.A.; Scarpello, K.E.; Morgan, N.G. The Imidazoline Site Involved in Control of Insulin Secretion: Characteristics That Distinguish It from I1- and I2-Sites. Br. J. Pharmacol. 1994, 112, 1065–1070. [Google Scholar] [CrossRef] [Green Version]
- Ernsberger, P.; Haxhiu, M.A. The I1-Imidazoline-Binding Site Is a Functional Receptor Mediating Vasodepression via the Ventral Medulla. Am. J. Physiol. 1997, 273, R1572–R1579. [Google Scholar] [CrossRef]
- Head, G.; Mayorov, D. Imidazoline Receptors, Novel Agents and Therapeutic Potential. Cardiovasc. Hematol. Agents Med. Chem. 2008, 4, 17–32. [Google Scholar] [CrossRef]
- Bousquet, P.; Hudson, A.; García-Sevilla, J.A.; Li, J.-X. Imidazoline Receptor System: The Past, the Present, and the Future. Pharmacol. Rev. 2020, 72, 50–79. [Google Scholar] [CrossRef]
- Li, J.-X. Imidazoline I2 Receptors: An Update. Pharmacol. Ther. 2017, 178, 48–56. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Z.-X.; Liu, Y.-F.; Xu, H.-Q.; Hou, S.-T.; Zheng, R.-Y. 2-BFI Ameliorates EAE-Induced Mouse Spinal Cord Damage: Effective Therapeutic Time Window and Possible Mechanisms. Brain Res. 2012, 1483, 13–19. [Google Scholar] [CrossRef]
- Zhu, Y.-B.; Xia, N.-G.; Zhang, Y.-T.; Wang, X.-S.; Liang, S.-S.; Yin, W.-Y.; Xu, H.-Q.; Hou, S.-T.; Zheng, R.-Y. Brain Protection Conferred by Long-Term Administration of 2-(2-Benzofuranyl)-2-Imidazoline against Experimental Autoimmune Encephalomyelitis. Neurochem. Res. 2015, 40, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Rui, Q.; Lin, X.; Li, D.; Liu, H.; Chen, G. 2-BFI Provides Neuroprotection against Inflammation and Necroptosis in a Rat Model of Traumatic Brain Injury. Front. Neurosci. 2019, 13, 674. [Google Scholar] [CrossRef]
- Siemian, J.N.; LaMacchia, Z.M.; Spreuer, V.; Tian, J.; Ignatowski, T.A.; Paez, P.M.; Zhang, Y.; Li, J.-X. The Imidazoline I2 Receptor Agonist 2-BFI Attenuates Hypersensitivity and Spinal Neuroinflammation in a Rat Model of Neuropathic Pain. Biochem. Pharmacol. 2018, 153, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Griñán-Ferré, C.; Vasilopoulou, F.; Abás, S.; Rodríguez-Arévalo, S.; Bagán, A.; Sureda, F.X.; Pérez, B.; Callado, L.F.; García-Sevilla, J.A.; García-Fuster, M.J.; et al. Behavioral and Cognitive Improvement Induced by Novel Imidazoline I2 Receptor Ligands in Female SAMP8 Mice. Neurotherapeutics 2019, 16, 416–431. [Google Scholar] [CrossRef] [Green Version]
- Vasilopoulou, F.; Bagan, A.; Rodriguez-Arevalo, S.; Escolano, C.; Griñán-Ferré, C.; Pallàs, M. Amelioration of BPSD-Like Phenotype and Cognitive Decline in SAMP8 Mice Model Accompanied by Molecular Changes after Treatment with I2-Imidazoline Receptor Ligand MCR5. Pharmaceutics 2020, 12, 475. [Google Scholar] [CrossRef] [PubMed]
- Vasilopoulou, F.; Griñán-Ferré, C.; Rodríguez-Arévalo, S.; Bagán, A.; Abás, S.; Escolano, C.; Pallàs, M. I2 Imidazoline Receptor Modulation Protects Aged SAMP8 Mice against Cognitive Decline by Suppressing the Calcineurin Pathway. GeroScience 2021, 43, 965–983. [Google Scholar] [CrossRef]
- Ruiz, J.; Martín, I.; Callado, L.F.; Meana, J.J.; Barturen, F.; García-Sevilla, J.A. Non-Adrenoceptor [3H]Idazoxan Binding Sites (I2-Imidazoline Sites) Are Increased in Postmortem Brain from Patients with Alzheimer’s Disease. Neurosci. Lett. 1993, 160, 109–112. [Google Scholar] [CrossRef]
- García-Sevilla, J.A.; Escribá, P.V.; Walzer, C.; Bouras, C.; Guimón, J. Imidazoline Receptor Proteins in Brains of Patients with Alzheimer’s Disease. Neurosci Lett. 1998, 247, 95–98. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Mielke, M.L.; Gómez-Isla, T.; Betensky, R.A.; Growdon, J.H.; Frosch, M.P.; Hyman, B.T. Reactive Glia Not Only Associates with Plaques but Also Parallels Tangles in Alzheimer’s Disease. Am. J. Pathol. 2011, 179, 1373–1384. [Google Scholar] [CrossRef]
- Ardura-Fabregat, A.; Boddeke, E.W.G.M.; Boza-Serrano, A.; Brioschi, S.; Castro-Gomez, S.; Ceyzériat, K.; Dansokho, C.; Dierkes, T.; Gelders, G.; Heneka, M.T.; et al. Targeting Neuroinflammation to Treat Alzheimer’s Disease. CNS Drugs 2017, 31, 1057–1082. [Google Scholar] [CrossRef] [Green Version]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal Beta-Amyloid Aggregates, Neurodegeneration, and Neuron Loss in Transgenic Mice with Five Familial Alzheimer’s Disease Mutations: Potential Factors in Amyloid Plaque Formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- García-Sevilla, J.A.; Alemany, R.; Olmos, G.; Menargues, A.; Obach, R. Chronic Imidazoline Drug Treatment Increases the Immunoreactivity of Glial Fibrillary Acidic Protein in Rat Brain. LSL 60101 as a Novel and Selective Ligand for I2-Imidazoline Receptors. Ann. N. Y. Acad. Sci. 1995, 763, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Arévalo, S.; Bagán, A.; Griñán-Ferré, C.; Vasilopoulou, F.; Pallàs, M.; Brocos-Mosquera, I.; Callado, L.F.; Loza, M.I.; Martínez, A.L.; Brea, J.; et al. Benzofuranyl-2-Imidazoles as Imidazoline I2 Receptor Ligands for Alzheimer’s Disease. Eur. J. Med. Chem. 2021, 222, 113540. [Google Scholar] [CrossRef] [PubMed]
- Alemany, R.; Olmos, G.; García-Sevilla, J.A. Chronic Treatment with Phenelzine and Other Irreversible Monoamine Oxidase Inhibitors Downregulates I2-Imidazoline Receptors in the Brain and Liver. Ann. N. Y. Acad. Sci. 1995, 763, 506–509. [Google Scholar] [CrossRef] [PubMed]
- Escribá, P.V.; Alemany, R.; Sastre, M.; Olmos, G.; Ozaita, A.; García-Sevilla, J.A. Pharmacological Modulation of Immunoreactive Imidazoline Receptor Proteins in Rat Brain: Relationship with Non-Adrenoceptor [3H]-Idazoxan Binding Sites. Br. J. Pharmacol. 1996, 118, 2029–2036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Blázquez, P.; Boronat, M.A.; Olmos, G.; García-Sevilla, J.A.; Garzón, J. Activation of I2-Imidazoline Receptors Enhances Supraspinal Morphine Analgesia in Mice: A Model to Detect Agonist and Antagonist Activities at These Receptors. Br. J. Pharmacol. 2000, 130, 146–152. [Google Scholar] [CrossRef] [Green Version]
- Mirzaei, N.; Mota, B.C.; Birch, A.M.; Davis, N.; Romero-Molina, C.; Katsouri, L.; Palmer, E.O.C.; Golbano, A.; Riggall, L.J.; Nagy, I.; et al. Imidazoline Ligand BU224 Reverses Cognitive Deficits, Reduces Microgliosis and Enhances Synaptic Connectivity in a Mouse Model of Alzheimer’s Disease. Br. J. Pharmacol. 2021, 178, 654–671. [Google Scholar] [CrossRef]
- Vasilopoulou, F.; Rodríguez-Arévalo, S.; Bagán, A.; Escolano, C.; Griñán-Ferré, C.; Pallàs, M. Disease-modifying Treatment with I 2 Imidazoline Receptor Ligand LSL60101 in an Alzheimer’s Disease Mouse Model: A Comparative Study with Donepezil. Br. J. Pharmacol. 2021, 178, bph.15478. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Tang, H.; Pan, Y.; Chen, J.; Li, Y.; Zhu, J. Gene Expression Profiles and Pathway Enrichment Analysis of Human Osteosarcoma Cells Exposed to Sorafenib. FEBS Open Bio 2018, 8, 860–867. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.-W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An Expanded Reference Database of Human and Mouse Transcriptional Regulatory Interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Nagae, T.; Araki, K. Cytokines and Cytokine Receptors Involved in the Pathogenesis of Alzheimers Disease. J. Clin. Cell Immunol. 2016, 7, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-ΚB Signaling in Inflammation. Sig. Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [Green Version]
- Jha, N.K.; Jha, S.K.; Kar, R.; Nand, P.; Swati, K.; Goswami, V.K. Nuclear Factor-Kappa β as a Therapeutic Target for Alzheimer’s Disease. J. Neurochem. 2019, 150, 113–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, F.; Edison, P. Neuroinflammation and Microglial Activation in Alzheimer Disease: Where Do We Go from Here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef]
- Passamonti, L.; Tsvetanov, K.A.; Jones, P.S.; Bevan-Jones, W.R.; Arnold, R.; Borchert, R.J.; Mak, E.; Su, L.; O’Brien, J.T.; Rowe, J.B. Neuroinflammation and Functional Connectivity in Alzheimer’s Disease: Interactive Influences on Cognitive Performance. J. Neurosci. 2019, 39, 7218–7226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gyengesi, E.; Münch, G. In Search of an Anti-Inflammatory Drug for Alzheimer Disease. Nat. Rev. Neurol. 2020, 16, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Abás, S.; Rodríguez-Arévalo, S.; Bagán, A.; Griñán-Ferré, C.; Vasilopoulou, F.; Brocos-Mosquera, I.; Muguruza, C.; Pérez, B.; Molins, E.; Luque, F.J.; et al. Bicyclic α-Iminophosphonates as High Affinity Imidazoline I2 Receptor Ligands for Alzheimer’s Disease. J. Med. Chem. 2020, 63, 3610–3633. [Google Scholar] [CrossRef]
- Fakhoury, M. Microglia and Astrocytes in Alzheimer’s Disease: Implications for Therapy. Curr. Neuropharmacol. 2018, 16, 508–518. [Google Scholar] [CrossRef]
- Martin, E.; Delarasse, C. Complex Role of Chemokine Mediators in Animal Models of Alzheimer’s Disease. Biomed. J. 2018, 41, 34–40. [Google Scholar] [CrossRef]
- Ryu, J.K.; Cho, T.; Choi, H.B.; Jantaratnotai, N.; McLarnon, J.G. Pharmacological Antagonism of Interleukin-8 Receptor CXCR2 Inhibits Inflammatory Reactivity and Is Neuroprotective in an Animal Model of Alzheimer’s Disease. J. Neuroinflamm. 2015, 12, 144. [Google Scholar] [CrossRef] [Green Version]
- Tan, J. Microglial Activation Resulting from CD40-CD40L Interaction After -Amyloid Stimulation. Science 1999, 286, 2352–2355. [Google Scholar] [CrossRef]
- Goldeck, D.; Larbi, A.; Pellicanó, M.; Alam, I.; Zerr, I.; Schmidt, C.; Fulop, T.; Pawelec, G. Enhanced Chemokine Receptor Expression on Leukocytes of Patients with Alzheimer’s Disease. PLoS ONE 2013, 8, e66664. [Google Scholar] [CrossRef] [Green Version]
- Boronat, M.A.; Olmos, G.; García-Sevilla, J.A. Attenuation of Tolerance to Opioid-Induced Antinociception and Protection against Morphine-Induced Decrease of Neurofilament Proteins by Idazoxan and Other I2-Imidazoline Ligands. Br. J. Pharmacol. 1998, 125, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Casanovas, A.; Olmos, G.; Ribera, J.; Boronat, M.A.; Esquerda, J.E.; García-Sevilla, J.A. Induction of Reactive Astrocytosis and Prevention of Motoneuron Cell Death by the I(2)-Imidazoline Receptor Ligand LSL 60101. Br. J. Pharmacol. 2000, 130, 1767–1776. [Google Scholar] [CrossRef] [Green Version]
- Menargues, A.; Cedó, M.; Artiga, O.; Obach, R.; García-Sevilla, J.A. Effects of the I2-Imidazoline Receptor Ligand LSL 60101 on Various Models of Anorexia in Ratsfn1. Ann. N. Y. Acad. Sci. 1995, 763, 494–496. [Google Scholar] [CrossRef]
- Ozaita, A.; Olmos, G.; Boronat, M.A.; Lizcano, J.M.; Unzeta, M.; García-Sevilla, J.A. Inhibition of Monoamine Oxidase A and B Activities by Imidazol(Ine)/Guanidine Drugs, Nature of the Interaction and Distinction from I2-Imidazoline Receptors in Rat Liver. Br. J. Pharmacol. 1997, 121, 901–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Xuan, Z.; Yang, M.; Wang, C.; Tao, T.; Wang, Q.; Cui, W. CSB6B Prevents β-Amyloid-Associated Neuroinflammation and Cognitive Impairments via Inhibiting NF-ΚB and NLRP3 in Microglia Cells. Int. Immunopharmacol. 2020, 81, 106263. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.V.; Kounatidis, I. Nuclear Factor-Kappa B and Alzheimer Disease, Unifying Genetic and Environmental Risk Factors from Cell to Humans. Front. Immunol. 2017, 8, 1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasilopoulou, F.; Escolano, C.; Pallàs, M.; Griñán-Ferré, C. Microarray Analysis Revealed Inflammatory Transcriptomic Changes after LSL60101 Treatment in 5XFAD Mice Model. Genes 2021, 12, 1315. https://doi.org/10.3390/genes12091315
Vasilopoulou F, Escolano C, Pallàs M, Griñán-Ferré C. Microarray Analysis Revealed Inflammatory Transcriptomic Changes after LSL60101 Treatment in 5XFAD Mice Model. Genes. 2021; 12(9):1315. https://doi.org/10.3390/genes12091315
Chicago/Turabian StyleVasilopoulou, Foteini, Carmen Escolano, Mercè Pallàs, and Christian Griñán-Ferré. 2021. "Microarray Analysis Revealed Inflammatory Transcriptomic Changes after LSL60101 Treatment in 5XFAD Mice Model" Genes 12, no. 9: 1315. https://doi.org/10.3390/genes12091315
APA StyleVasilopoulou, F., Escolano, C., Pallàs, M., & Griñán-Ferré, C. (2021). Microarray Analysis Revealed Inflammatory Transcriptomic Changes after LSL60101 Treatment in 5XFAD Mice Model. Genes, 12(9), 1315. https://doi.org/10.3390/genes12091315







