Integrated Bioinformatics Analysis Reveals Marker Genes and Potential Therapeutic Targets for Pulmonary Arterial Hypertension
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Selection
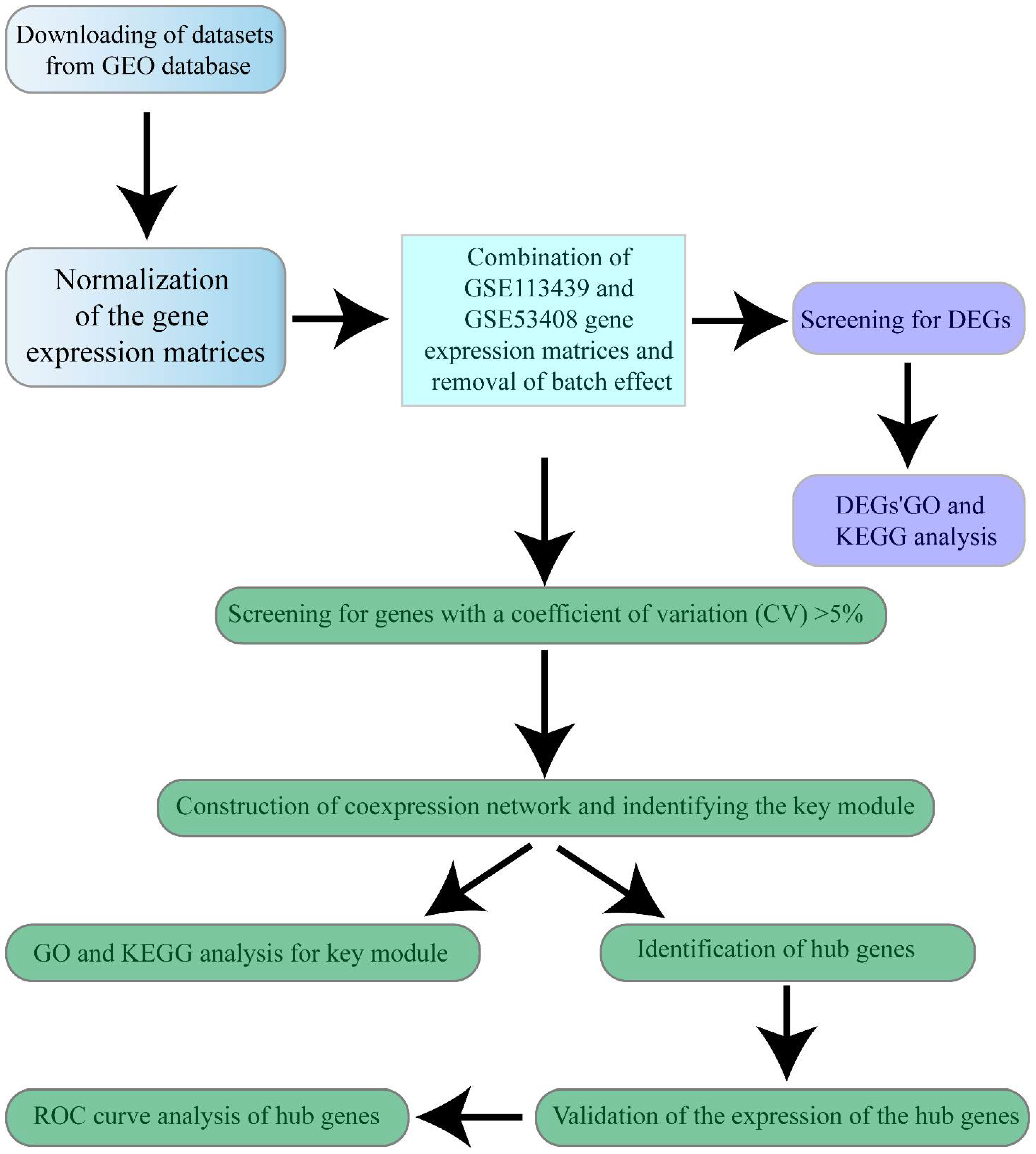
2.2. Data Preprocessing and DEGs Screening
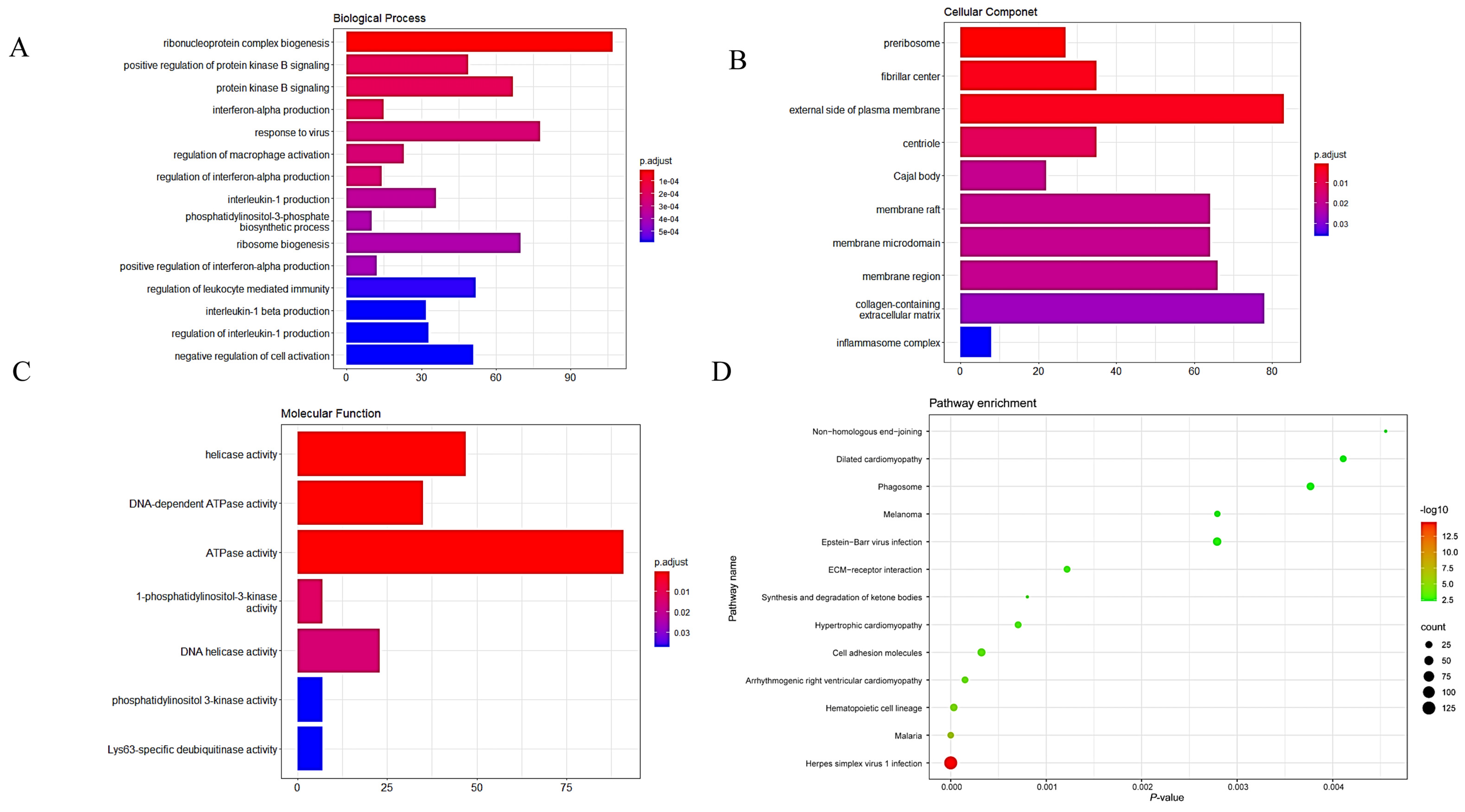
2.3. GO and KEGG Functional Enrichment Analyses
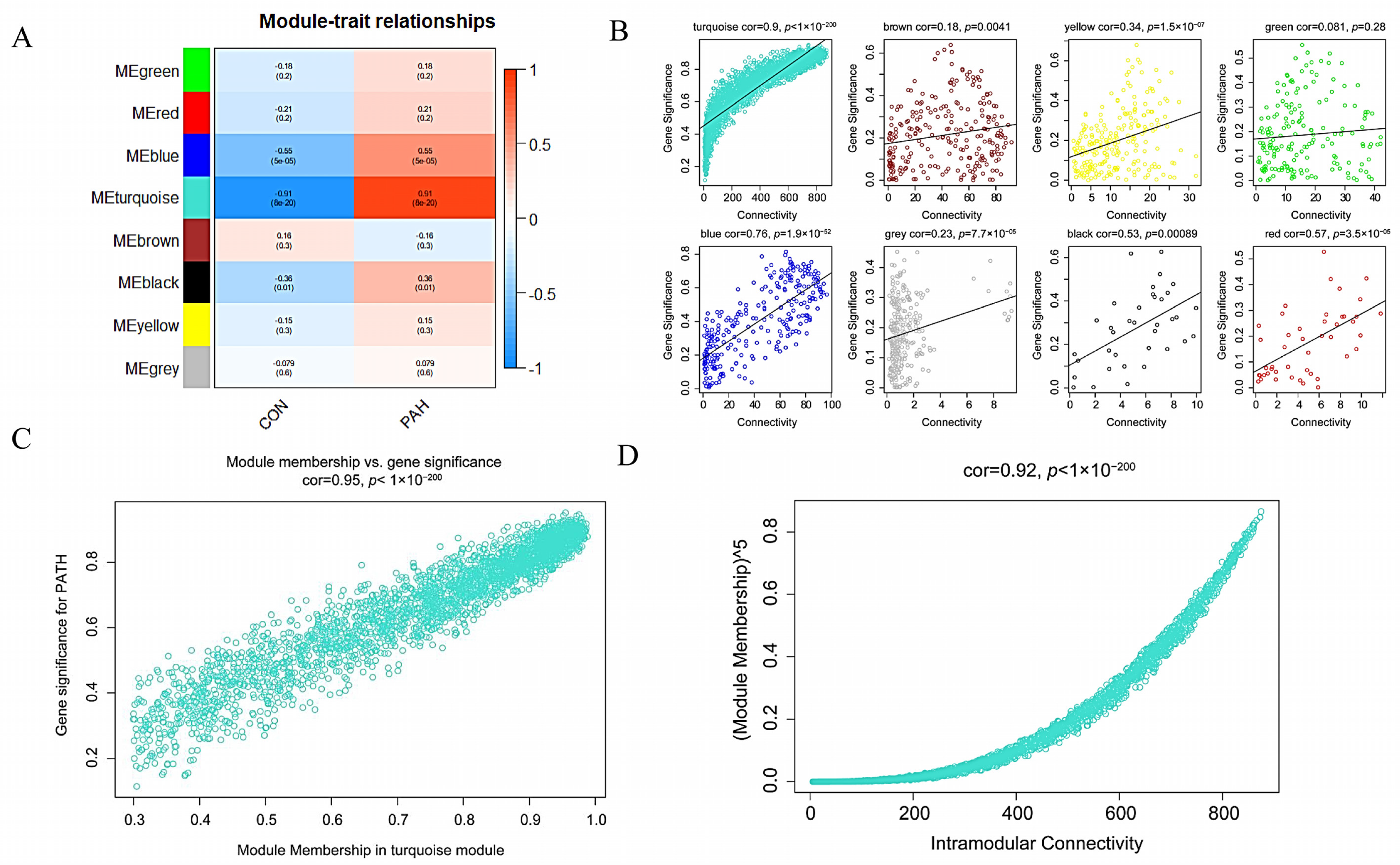
2.4. Co-Expression Network Analysis
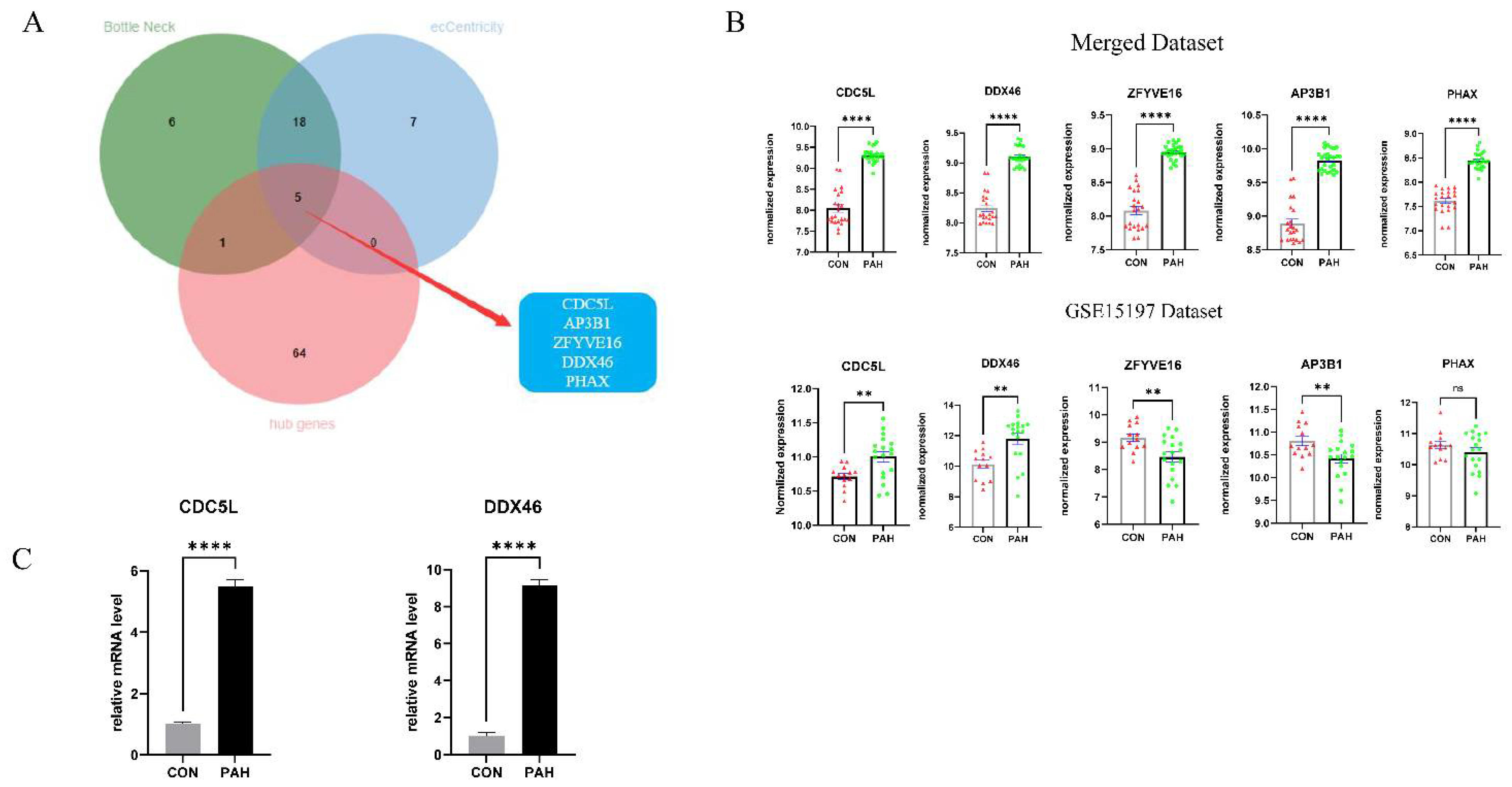
2.5. Identification of Candidate Marker Genes
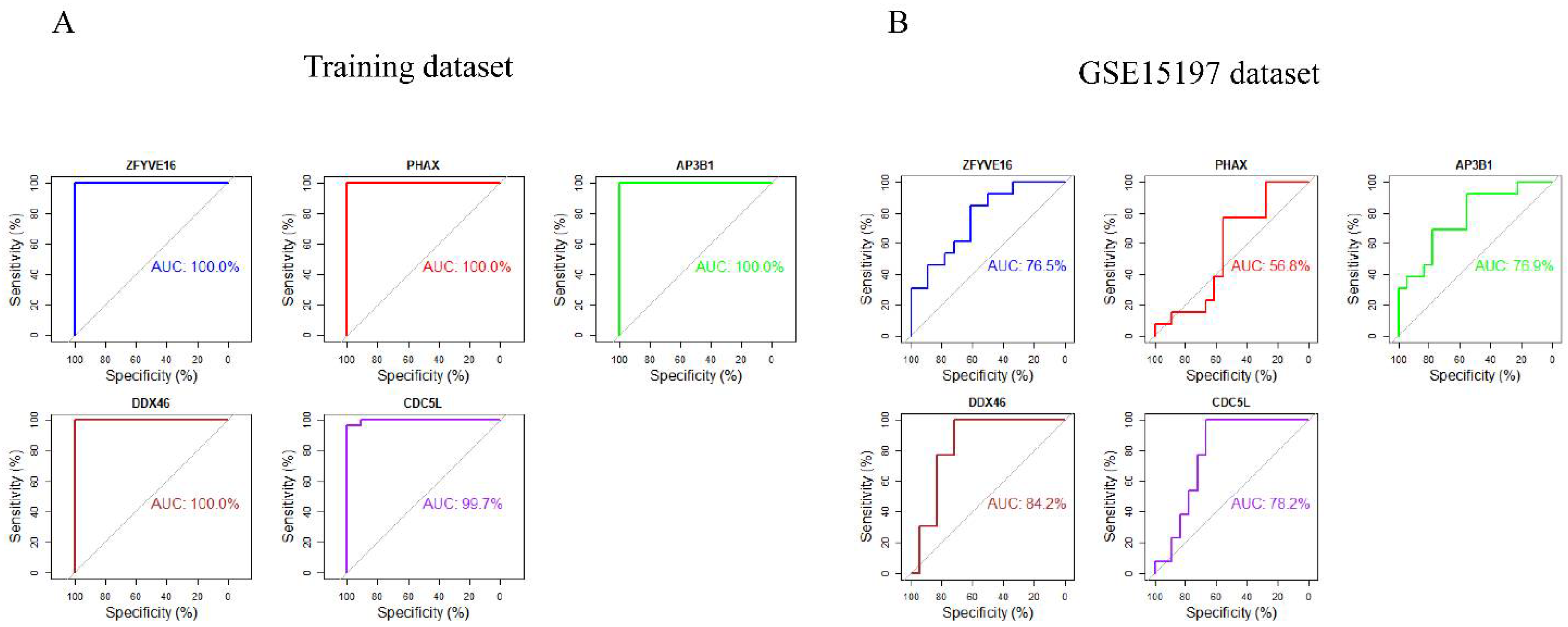
2.6. Validation of Candidate Marker Genes and ROC Curve Analyses
2.7. PAH Model and qRT-PCR
2.8. Statistical Analysis
3. Results
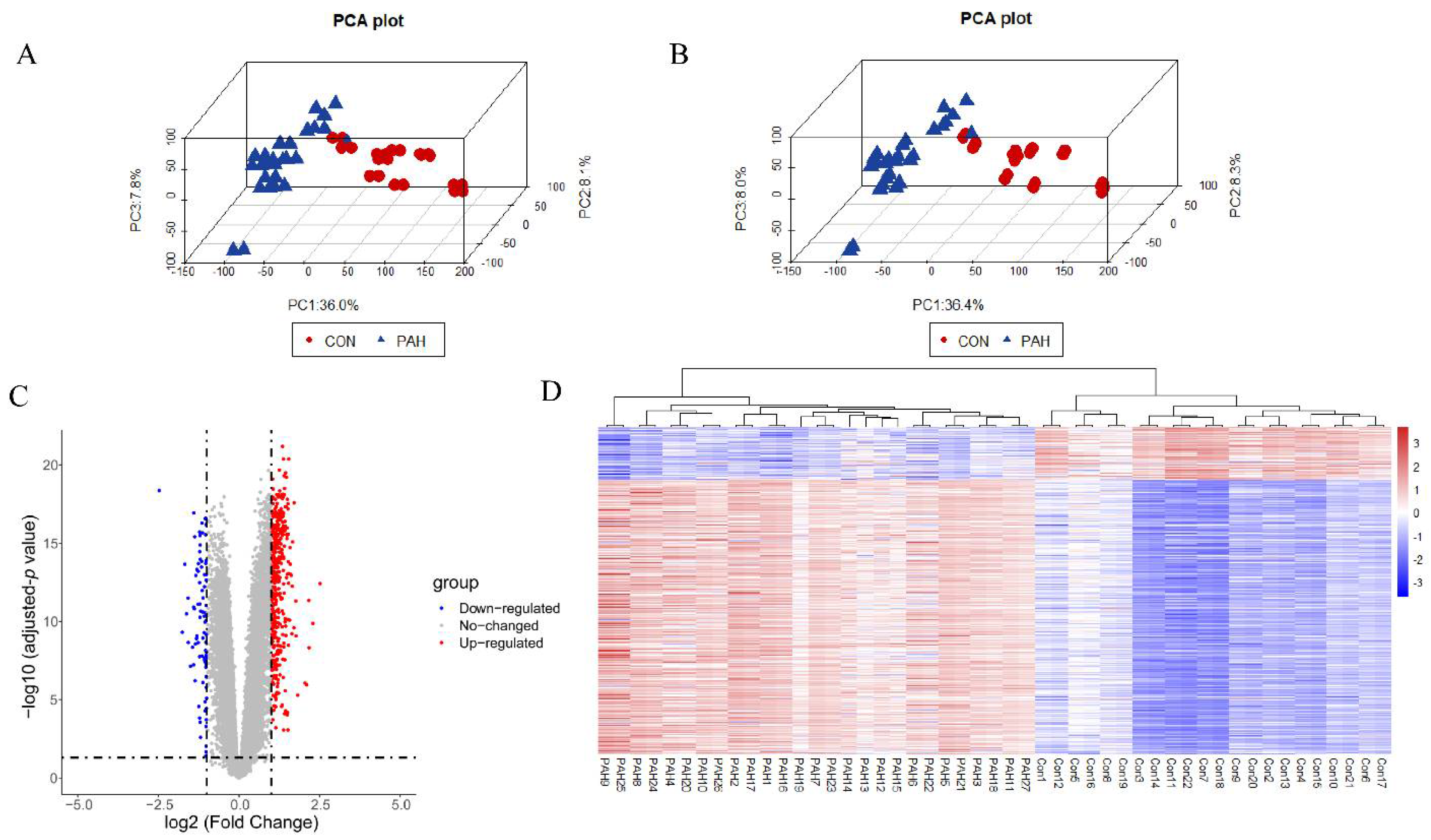
3.1. Data Preprocessing and DEGs Screening
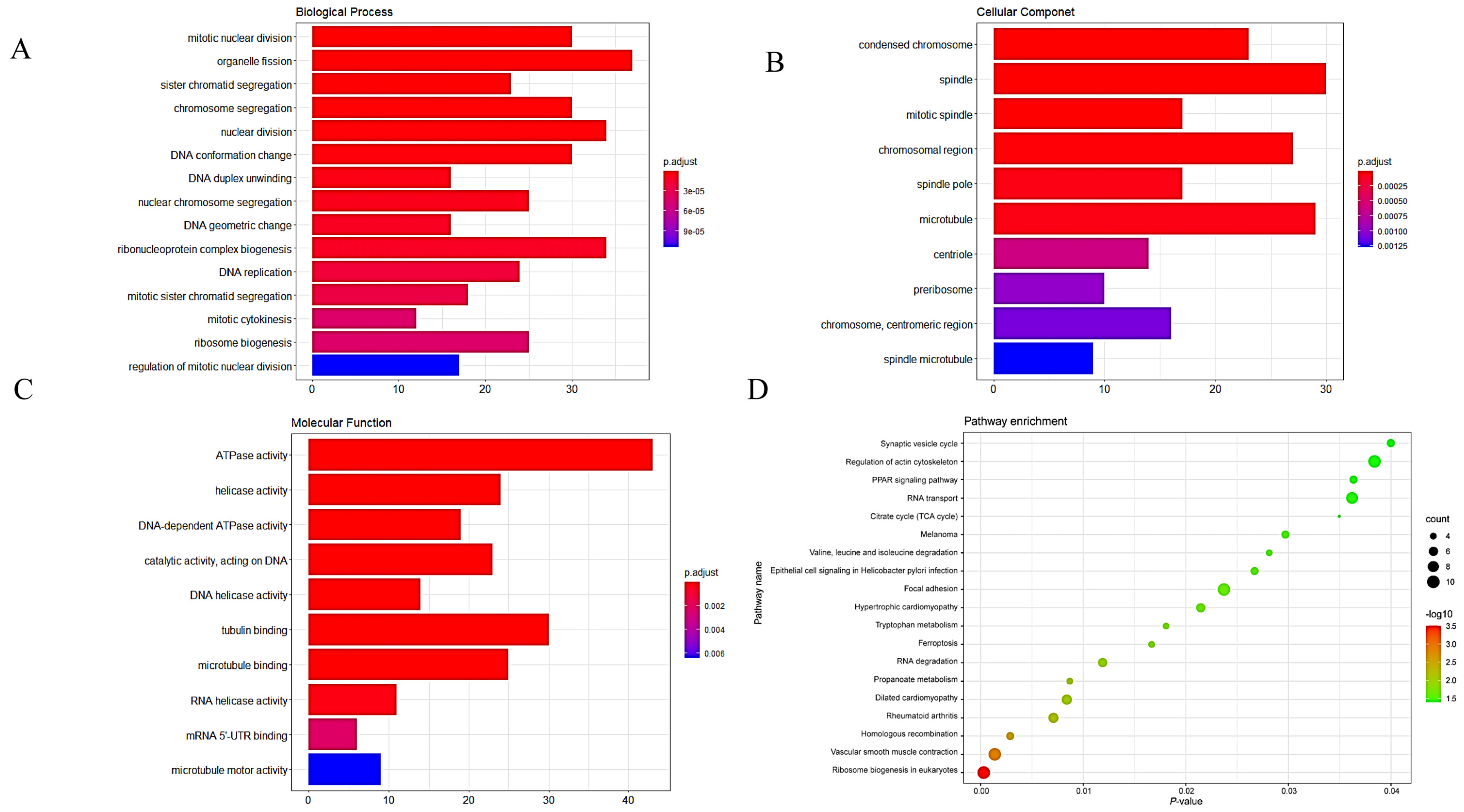
3.2. GO and KEGG Analyses of DEGs
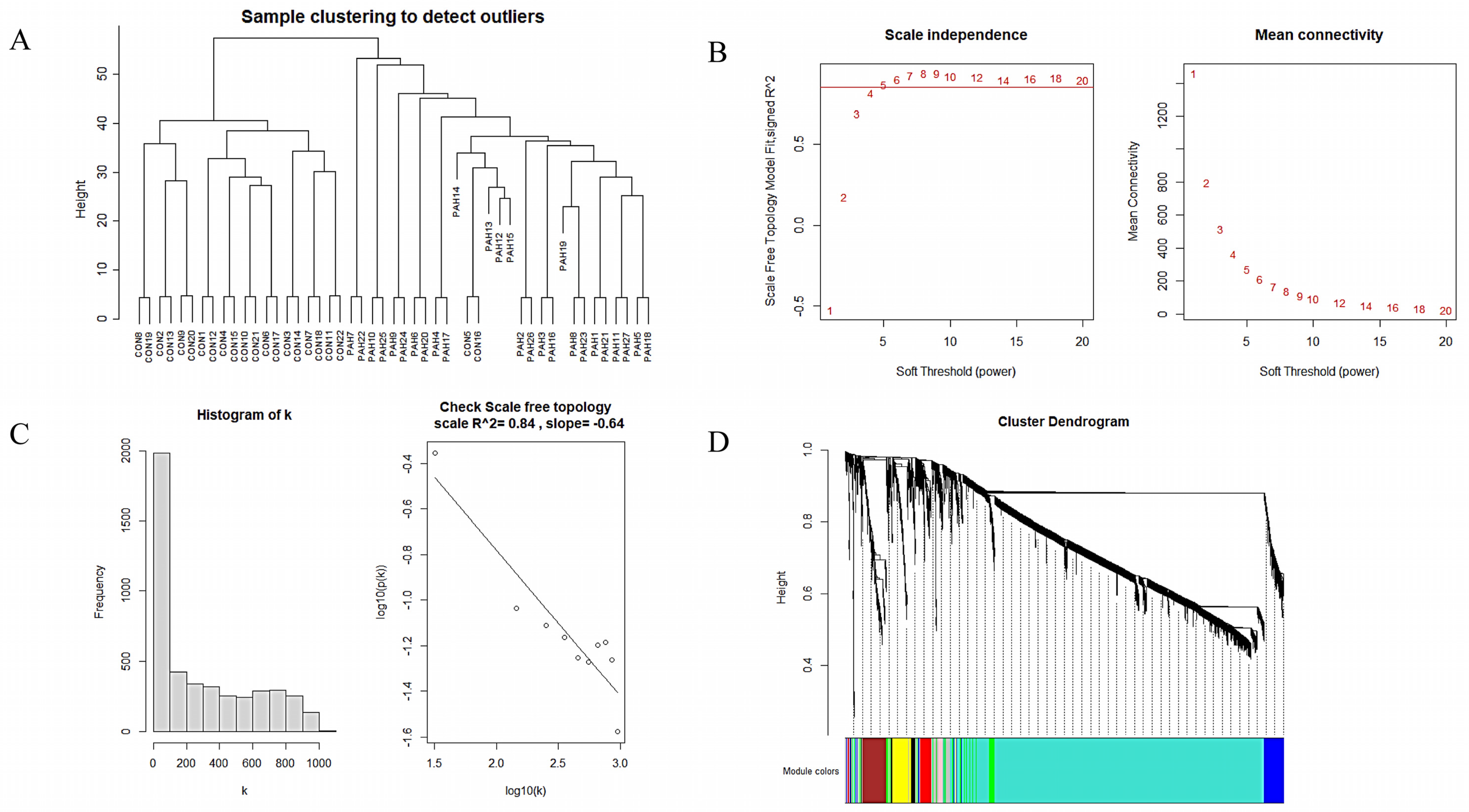
3.3. Construction of WGCNA Network and Identification of the Key Module
3.4. GO and KEGG Analyses of the Key Module
3.5. Identification of Hub Genes
3.6. Validation of Candidate Marker Genes
3.7. ROC Curve Analyses of Hub Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zolty, R. Pulmonary arterial hypertension specific therapy: The old and the new. Pharmacol. Ther. 2020, 214, 107576. [Google Scholar] [CrossRef] [PubMed]
- Humbert, M.; Sitbon, O.; Chaouat, A.; Bertocchi, M.; Habib, G.; Gressin, V.; Yaici, A.; Weitzenblum, E.; Cordier, J.-F.; Chabot, F.; et al. Pulmonary Arterial Hypertension in France. Am. J. Respir. Crit. Care Med. 2006, 173, 1023–1030. [Google Scholar] [CrossRef] [Green Version]
- Bhogal, S.; Khraisha, O.; Al Madani, M.; Treece, J.; Baumrucker, S.; Paul, T.K. Sildenafil for Pulmonary Arterial Hypertension. Am. J. Ther. 2019, 26, e520–e526. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhai, Z.; Huang, K.; Xie, W.; Wan, J.; Wang, C. Bosentan therapy for pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension: A systemic review and meta-analysis. Clin. Respir. J. 2018, 12, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, A.; Kadry, H.; Alobaida, A.; Ahsan, F. Newer approaches and novel drugs for inhalational therapy for pulmonary arterial hypertension. Expert Opin. Drug Deliv. 2020, 17, 439–461. [Google Scholar] [CrossRef]
- Toxvig, A.K.; Wehland, M.; Grimm, D.; Infanger, M.; Krüger, M. A focus on riociguat in the treatment of pulmonary arterial hypertension. Basic Clin. Pharmacol. Toxicol. 2019, 125, 202–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benza, R.L.; Miller, D.; Barst, R.J.; Badesch, D.B.; Frost, A.E.; McGoon, M.D. An Evaluation of Long-term Survival From Time of Diagnosis in Pulmonary Arterial Hypertension From the REVEAL Registry. Chest 2012, 142, 448–456. [Google Scholar] [CrossRef]
- Thenappan, T.; Ormiston, M.; Ryan, J.J.; Archer, S.L. Pulmonary arterial hypertension: Pathogenesis and clinical management. BMJ 2018, 360, j5492. [Google Scholar] [CrossRef]
- Van’T Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [Green Version]
- Buyse, M.; Loi, S.; Veer, L.V.; Viale, G.; Delorenzi, M.; Glas, A.; D’Assignies, M.S.; Bergh, J.; Lidereau, R.; Ellis, P.; et al. Validation and Clinical Utility of a 70-Gene Prognostic Signature for Women With Node-Negative Breast Cancer. J. Natl. Cancer Inst. 2006, 98, 1183–1192. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Tian, Z.; He, W.; Tang, J.; Liao, X.; Yang, Q.; Wu, Y.; Wu, G. Identification of Important Modules and Biomarkers in Breast Cancer Based on WGCNA. OncoTargets Ther. 2020, 13, 6805–6817. [Google Scholar] [CrossRef]
- Luo, Y.; Coskun, V.; Liang, A.; Yu, J.; Cheng, L.; Ge, W.; Shi, Z.; Zhang, K.; Li, C.; Cui, Y.; et al. Single-Cell Transcriptome Analyses Reveal Signals to Activate Dormant Neural Stem Cells. Cell 2015, 161, 1175–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, X.; Zhang, J.; Zhang, L.; Hou, Y.; Pu, S.; Chu, A.; Bai, M.; Zhang, Z. Weighted Gene Co-Expression Network Analysis Identifies Critical Genes in the Development of Heart Failure After Acute Myocardial Infarction. Front. Genet. 2019, 10, 1214. [Google Scholar] [CrossRef] [Green Version]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Aff—Analysis of Affymetrix GeneChip data at the probe level. Bioinforma 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Richard, A.C.; Lyons, P.A.; Peters, J.E.; Biasci, D.; Flint, S.M.; Lee, J.C.; McKinney, E.F.; Siegel, R.M.; Smith, K.G.C. Comparison of gene expression microarray data with count-based RNA measurements informs microarray interpretation. BMC Genom. 2014, 15, 649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.S.; Leek, J.T.; Favorov, A.V.; Considine, M.; Xia, X.; Chavan, S.; Chung, C.H.; Fertig, E.J. Preserving biological heterogeneity with a permuted surrogate variable analysis for genomics batch correction. Bioinforma 2014, 30, 2757–2763. [Google Scholar] [CrossRef] [Green Version]
- Ligges, U.; Mächler, M. Scatterplot3d—An R Package for Visualizing Multivariate Data. J. Stat. Softw. 2003, 8, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [Green Version]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Muller, M.J. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Chapman, M.E.; Wideman, R.F. Pulmonary Wedge Pressures Confirm Pulmonary Hypertension in Broilers Is Initiated by an Excessive Pulmonary Arterial (Precapillary) Resistance. Poult. Sci. 2001, 80, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Eichstaedt, C.A.; Verweyen, J.; Halank, M.; Benjamin, N.; Fischer, C.; Mayer, E.; Guth, S.; Wiedenroth, C.B.; Egenlauf, B.; Harutyunova, S.; et al. Myeloproliferative Diseases as Possible Risk Factor for Development of Chronic Thromboembolic Pulmonary Hypertension—A Genetic Study. Int. J. Mol. Sci. 2020, 21, 3339. [Google Scholar] [CrossRef]
- Wang, K.; Zhao, S.; Liu, B.; Zhang, Q.; Li, Y.; Liu, J.; Shen, Y.; Ding, X.; Lin, J.; Wu, Y.; et al. Perturbations of BMP/TGF-β and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM). J. Med. Genet. 2018, 55, 675–684. [Google Scholar] [CrossRef]
- Conidi, A.; Berghe, V.V.D.; Huylebroeck, D. Aptamers and Their Potential to Selectively Target Aspects of EGF, Wnt/β-Catenin and TGFβ–Smad Family Signaling. Int. J. Mol. Sci. 2013, 14, 6690–6719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Eichstaedt, C.A.; Viales, R.R.; Benjamin, N.; Harutyunova, S.; Fischer, C.; Grünig, E.; Hinderhofer, K. Identification of genetic defects in pulmonary arterial hypertension by a new gene panel diagnostic tool. Clin. Sci. 2016, 130, 2043–2052. [Google Scholar] [CrossRef]
- Nishikawa, T.; Okamura, K.; Moriyama, M.; Watanabe, K.; Ibusuki, A.; Sameshima, S.; Masamoto, I.; Yamazaki, I.; Tanita, K.; Kanekura, T.; et al. Novel AP3B1 compound heterozygous mutations in a Japanese patient with Hermansky–Pudlak syndrome type. J. Dermatol. 2020, 47, 185–189. [Google Scholar] [CrossRef]
- Summer, R.; Krishna, R.; Schriner, D.; Cuevas-Mora, K.; Sales, D.; Para, R.; Roman, J.; Nieweld, C.; Gochuico, B.R.; Romero, F. Matrix metalloproteinase activity in the lung is increased in Hermansky-Pudlak syndrome. Orphanet J. Rare Dis. 2019, 14, 162. [Google Scholar] [CrossRef] [Green Version]
- Mourão, A.; Varrot, A.; Mackereth, C.D.; Cusack, S.; Sattler, M. Structure and RNA recognition by the snRNA and snoRNA transport factor PHAX. RNA 2010, 16, 1205–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naito, A.; Hiwasa, T.; Tanabe, N.; Sanada, T.J.; Sugiura, T.; Shigeta, A.; Terada, J.; Takizawa, H.; Kashiwado, K.; Sakao, S.; et al. Elevated levels of autoantibodies against EXD2 and PHAX in the sera of patients with chronic thromboembolic pulmonary hypertension. PLoS ONE 2019, 14, e0211377. [Google Scholar] [CrossRef] [PubMed]
- Machitani, M.; Taniguchi, I.; McCloskey, A.; Suzuki, T.; Ohno, M. The RNA transport factor PHAX is required for proper histone H2AX expression and DNA damage response. RNA 2020, 26, 1716–1725. [Google Scholar] [CrossRef]
- Sharma, S.; Aldred, M.A. DNA Damage and Repair in Pulmonary Arterial Hypertension. Genes 2020, 11, 1224. [Google Scholar] [CrossRef] [PubMed]
- Grote, M.; Wolf, E.; Will, C.L.; Lemm, I.; Agafonov, D.E.; Schomburg, A.; Fischle, W.; Urlaub, H.; Lührmann, R. Molecular Architecture of the Human Prp19/CDC5L Complex. Mol. Cell. Biol. 2010, 30, 2105–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Mao, W.; Wang, L.; Liu, M.; Zhang, W.; Wu, Y.; Zhang, J.; Mao, S.; Geng, J.; Yao, X. Depletion of CDC5L inhibits bladder cancer tumorigenesis. J. Cancer 2020, 11, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, X.; Song, W.; Xu, H.; Huang, R.; Wang, Y.; Zhao, W.; Xiao, Z.; Yang, X. Oncogenic Properties of NEAT1 in Prostate Cancer Cells Depend on the CDC5L–AGRN Transcriptional Regulation Circuit. Cancer Res. 2018, 78, 4138–4149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhen, L.; Ning, G.; Wu, L.; Zheng, Y.; Yang, F.; Chen, T.; Xu, W.; Liu, Y.; Xie, C.; Peng, L. Prognostic value of aberrantly expressed methylation genes in human hepatocellular carcinoma. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Zhou, R.; Zhang, J.-T.; Chen, C.; Xu, Z.-H.; Lv, X.-B.; Ye, L.; Yu, B.-T. Identification of CDC5L as bridge gene between chronic obstructive pulmonary disease and lung adenocarcinoma. Epigenomics 2020, 12, 1515–1529. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, Z.; Zhang, D.; Fu, L.; Wan, R.; Hong, K. Comparative Transcriptional Analysis of Pulmonary Arterial Hypertension Associated With Three Different Diseases. Front. Cell Dev. Biol. 2021, 9, 672159. [Google Scholar] [CrossRef]
- Zheng, Q.; Hou, J.; Zhou, Y.; Qingliang, Z.; Cao, X. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017, 18, 1094–1103. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Urlaub, H.; Achsel, T.; Gentzel, M.; Wilm, M.; Lührmann, R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002, 21, 4978–4988. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, R.; Hozumi, S.; Higashijima, S.-I.; Kikuchi, Y. Ddx46 Is Required for Multi-Lineage Differentiation of Hematopoietic Stem Cells in Zebrafish. Stem Cells Dev. 2013, 22, 2532–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Li, Y.-M.; He, W.-T.; Chen, H.; Zhu, H.-W.; Liu, T.; Zhang, J.-H.; Song, T.-N.; Zhou, Y.-L. Knockdown of DDX46 inhibits proliferation and induces apoptosis in esophageal squamous cell carcinoma cells. Oncol. Rep. 2016, 36, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Admoni-Elisha, L.; Nakdimon, I.; Shteinfer, A.; Prezma, T.; Arif, T.; Arbel, N.; Melkov, A.; Zelichov, O.; Levi, I.; Shoshan-Barmatz, V. Novel Biomarker Proteins in Chronic Lymphocytic Leukemia: Impact on Diagnosis, Prognosis and Treatment. PLoS ONE 2016, 11, e0148500. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Ma, Y.; Huang, P.; Du, A.; Yang, X.; Zhang, S.; Xing, C.; Liu, F.; Cao, J. Lentiviral DDX46 knockdown inhibits growth and induces apoptosis in human colorectal cancer cells. Gene 2015, 560, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Jin, H.-J.; Zhang, D.; Gao, L. DDX46 silencing inhibits cell proliferation by activating apoptosis and autophagy in cutaneous squamous cell carcinoma. Mol. Med. Rep. 2020, 22, 4236–4242. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, D.; Li, G.; Wang, X. Knockdown of DDX46 Inhibits the Invasion and Tumorigenesis in Osteosarcoma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 417–425. [Google Scholar] [CrossRef]
- Zhang, C.; Kuang, M.; Li, M.; Feng, L.; Zhang, K.; Cheng, S. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci. Rep. 2016, 6, 34508. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Xu, M.; Zhong, W.; Hu, Y.; Wang, G. Knockdown of DDX46 suppresses the proliferation and invasion of gastric cancer through inactivating Akt/GSK-3β/β-catenin pathway. Exp. Cell Res. 2021, 399, 112448. [Google Scholar] [CrossRef]
- Li, S.; Zhai, C.; Shi, W.; Feng, W.; Xie, X.; Pan, Y.; Wang, J.; Yan, X.; Chai, L.; Wang, Q.; et al. Leukotriene B4 induces proliferation of rat pulmonary arterial smooth muscle cells via modulating GSK-3β/β-catenin pathway. Eur. J. Pharmacol. 2020, 867, 172823. [Google Scholar] [CrossRef]
- Yu, R.H.; Wang, L.M.; Hu, X.H. MiR-135a inhibitor alleviates pulmonary arterial hypertension through beta-Catenin/GSK-3beta signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9574–9581. [Google Scholar] [CrossRef] [PubMed]
- Sitbon, O.; Channick, R.; Chin, K.; Frey, A.; Gaine, S.; Galiè, N.; Ghofrani, A.; Hoeper, M.; Lang, I.M.; Preiss, R.; et al. Selexipag for the Treatment of Pulmonary Arterial Hypertension. N. Engl. J. Med. 2015, 373, 2522–2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Roman-Campos, D.; Austin, E.D.; Eyries, M.; Sampson, K.S.; Soubrier, F.; Germain, M.; Trégouët, D.-A.; Borczuk, A.; Rosenzweig, E.B.; et al. A Novel Channelopathy in Pulmonary Arterial Hypertension. N. Engl. J. Med. 2013, 369, 351–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.-J.; Lian, T.-Y.; Jiang, X.; Liu, S.-F.; Li, S.-Q.; Jiang, R.; Wu, W.-H.; Ye, J.; Cheng, C.-Y.; Du, Y.; et al. Germline BMP9 mutation causes idiopathic pulmonary arterial hypertension. Eur. Respir. J. 2019, 53, 1801609. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.W.; Girerd, B.; Montani, D.; Wang, X.-J.; Galiè, N.; Austin, E.D.; Elliott, G.; Asano, K.; Grünig, E.; Yan, Y.; et al. BMPR2 mutations and survival in pulmonary arterial hypertension: An individual participant data meta-analysis. Lancet Respir. Med. 2016, 4, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Rol, N.; Kurakula, K.B.; Happé, C.; Bogaard, H.J.; Goumans, M.-J. TGF-β and BMPR2 Signaling in PAH: Two Black Sheep in One Family. Int. J. Mol. Sci. 2018, 19, 2585. [Google Scholar] [CrossRef] [Green Version]
- Drake, K.M.; Dunmore, B.J.; McNelly, L.N.; Morrell, N.W.; Aldred, M.A. Correction of NonsenseBMPR2andSMAD9Mutations by Ataluren in Pulmonary Arterial Hypertension. Am. J. Respir. Cell Mol. Biol. 2013, 49, 403–409. [Google Scholar] [CrossRef] [Green Version]
- Hamaguchi, Y.; Matsushita, T.; Hasegawa, M.; Ueda-Hayakawa, I.; Sato, S.; Takehara, K.; Fujimoto, M. High incidence of pulmonary arterial hypertension in systemic sclerosis patients with anti-centriole autoantibodies. Mod. Rheumatol. 2015, 25, 798–801. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Fork, C.; Josipovic, I.; Richter, F.M.; Preussner, J.; Hu, J.; Miller, M.J.; Epah, J.; Hofmann, P.; Günther, S.; et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation 2017, 136, 65–79. [Google Scholar] [CrossRef]
- Lampron, M.-C.; Vitry, G.; Nadeau, V.; Grobs, Y.; Paradis, R.; Samson, N.; Tremblay, È.; Boucherat, O.; Meloche, J.; Bonnet, S.; et al. PIM1 (Moloney Murine Leukemia Provirus Integration Site) Inhibition Decreases the Nonhomologous End-Joining DNA Damage Repair Signaling Pathway in Pulmonary Hypertension. Arter. Thromb. Vasc. Biol. 2020, 40, 783–801. [Google Scholar] [CrossRef]
- Covella, M.; Rowin, E.J.; Hill, N.S.; Preston, I.R.; Milan, A.; Opotowsky, A.R.; Maron, B.J.; Maron, M.S.; Maron, B.A. Mechanism of Progressive Heart Failure and Significance of Pulmonary Hypertension in Obstructive Hypertrophic Cardiomyopathy. Circ. Heart Fail. 2017, 10, e003689. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.X.; Kwong, C.; Swaminathan, A.; Gohil, A.; Crawford, M.H. Clinical Characteristics and Outcome of Methamphetamine-Associated Pulmonary Arterial Hypertension and Dilated Cardiomyopathy. JACC Heart Fail. 2018, 6, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Sargin, G.; Senturk, T.; Cildag, S. Systemic sclerosis and malignancy. Int. J. Rheum. Dis. 2018, 21, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Wideman, R.F. Pathophysiology of heart/lung disorders: Pulmonary hypertension syndrome in broiler chickens. World’s Poult. Sci. J. 2001, 57, 289–307. [Google Scholar] [CrossRef]
- Julian, R.J. Physiological, management and environmental triggers of the ascites syndrome: A review. Avian Pathol. 2000, 29, 519–527. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, A.; He, J.; Zhang, Z.; Jiang, S.; Gao, Y.; Pan, Y.; Wang, H.; Zhuang, L. Integrated Bioinformatics Analysis Reveals Marker Genes and Potential Therapeutic Targets for Pulmonary Arterial Hypertension. Genes 2021, 12, 1339. https://doi.org/10.3390/genes12091339
Li A, He J, Zhang Z, Jiang S, Gao Y, Pan Y, Wang H, Zhuang L. Integrated Bioinformatics Analysis Reveals Marker Genes and Potential Therapeutic Targets for Pulmonary Arterial Hypertension. Genes. 2021; 12(9):1339. https://doi.org/10.3390/genes12091339
Chicago/Turabian StyleLi, Aoqi, Jin He, Zhe Zhang, Sibo Jiang, Yun Gao, Yuchun Pan, Huanan Wang, and Lenan Zhuang. 2021. "Integrated Bioinformatics Analysis Reveals Marker Genes and Potential Therapeutic Targets for Pulmonary Arterial Hypertension" Genes 12, no. 9: 1339. https://doi.org/10.3390/genes12091339
APA StyleLi, A., He, J., Zhang, Z., Jiang, S., Gao, Y., Pan, Y., Wang, H., & Zhuang, L. (2021). Integrated Bioinformatics Analysis Reveals Marker Genes and Potential Therapeutic Targets for Pulmonary Arterial Hypertension. Genes, 12(9), 1339. https://doi.org/10.3390/genes12091339







