Development and Validation of KASP Assays for the Genotyping of Racing Performance-Associated Single Nucleotide Polymorphisms in Pigeons
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Sample Collection
2.2. PCR-RFLP Genotyping
2.3. KASP Assay
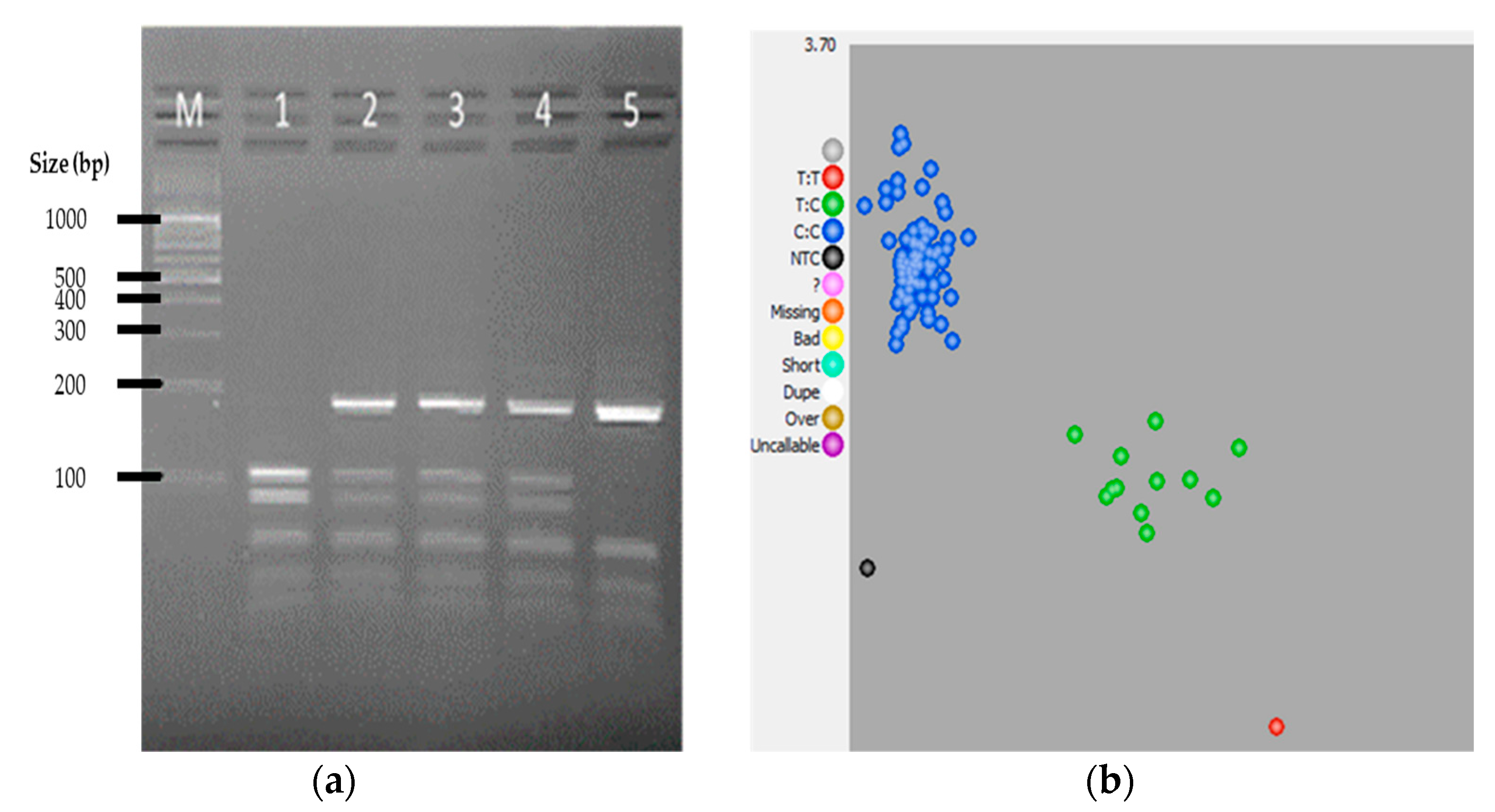
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dybus, A.; Kmieć, M. PCR-RFLPs within the Iactate dehydrogenase (LDH-A) gene of the domestic pigeon (Columba livia var. domestica). J. Appl. Genet. 2002, 43, 501–504. [Google Scholar] [PubMed]
- Jędrzejczak-Silicka, M.; Yu-Hsiang, Y.; Yeong-Hsiang, C.; Dybus, A. the Influence of Ldha Gene Polymorphism on Relative Level of Its Expression in Racing Pigeons. Acta Sci. Pol. Zootech. 2018, 17, 9–16. [Google Scholar] [CrossRef]
- Jerolmack, C. Animal archeology: Domestic pigeons and the nature-culture dialectic. Qual. Sociol. Rev. 2007, III, 74–95. [Google Scholar]
- Traxler, B.; Brem, G.; Muller, M.; Achmann, R. Polymorphic DNA microsatellites in the domestic pigeon, Columba livia var. domestica. Mol. Ecol. 2000, 9, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Abolnik, C. A current review of avian influenza in pigeons and doves (Columbidae). Vet. Microbiol. 2014, 170, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Proskura, W.S.; Cichoń, D.; Grzesiak, W.; Zaborski, D.; Sell-Kubiak, E.; Cheng, Y.H.; Dybus, A. Single nucleotide polymorphism in the LDHA gene as a potential marker for the racing performance of pigeons. J. Poult. Sci. 2014, 51, 364–368. [Google Scholar] [CrossRef]
- Proskura, W.S.; Dybus, A.; Lukaszewicz, A.; Hardziejewicz, E.; Pawlina, E. The single nucleotide polymorphisms in lactate dehydrogenase-a (LDHA) and feather keratin (F-KER) genes and racing performance of domestic pigeon. Zesz. Nauk. UP Wroc. Biol. Hod. Zwierz. 2015, LXXVI, 37–42. [Google Scholar]
- Ramadan, S.; Miyake, T.; Yamaura, J.; Inoue-Murayama, M. LDHA gene is associated with pigeon survivability during racing competitions. PLoS ONE 2018, 13, 1–9. [Google Scholar] [CrossRef]
- Dybus, A.; Pijanka, J.; Cheng, Y.H.; Sheen, F.; Grzesiak, W.; Muszyńska, M. Polymorphism within the LDHA gene in the homing and non-homing pigeons. J. Appl. Genet. 2006, 47, 63–66. [Google Scholar] [CrossRef]
- Dybus, A.; Proskura, W.S.; Sadkowski, S.; Pawlina, E. A single nucleotide polymorphism in exon 3 of the myostatin gene in different breeds of domestic pigeon (Columba livia var. domestica). Vet. Med. (Praha) 2013, 58, 32–38. [Google Scholar] [CrossRef]
- Firyal, S.; Awan, A.R.; Yaqub, T.; Anjum, A.A.; Asif, M.; Tayyab, M. Molecular classification of Pakistani domestic pigeon using cytochrome b gene. Pak. Vet. J. 2014, 34, 254–256. [Google Scholar]
- Proskura, W.S.; Lukaszewicz, A.; Dzierzba, E.; Cichon, D.; Zaborski, D.; Grzesiak, W.; Dybus, A. The Cys83Gly amino acid substitution in feather keratin is associated with pigeon performance in long-distance races. Vet. Med. (Praha) 2017, 62, 221–225. [Google Scholar] [CrossRef]
- Ramadan, S.; Yamaura, J.; Miyake, T.; Inoue-Murayama, M. DNA polymorphism within LDH-A gene in pigeon (Columba livia). J. Poult. Sci. 2013, 50, 194–197. [Google Scholar] [CrossRef][Green Version]
- Santos, H.M.; Chen, C.C.; Tsai, C.Y.; Hsish, Y.C.; Chung, F.C.; Tyan, Y.C.; Tayo, L.L.; Chuang, K.P. Influence of pigeon interferon alpha (PiIFN-α) on pigeon circovirus (PiCV) replication and cytokine expression in Columba livia. Vet. Microbiol. 2020, 242, 108591. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.M.; Tsai, C.Y.; Catulin, G.E.M.; Trangia, K.C.G.; Tayo, L.L.; Liu, H.J.; Chuang, K.P. Common bacterial, viral, and parasitic diseases in pigeons (Columba livia): A review of diagnostic and treatment strategies. Vet. Microbiol. 2020, 247, 108779. [Google Scholar] [CrossRef]
- Chaplin, S.B.; Munson, M.M.; Knuth, S.T. The effect of exercise and restraint on pectoral muscle metabolism in pigeons. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 1997, 167, 197–203. [Google Scholar] [CrossRef]
- Dybus, A.; Proskura, W.S.; Pawlina, E.; Nowak, B. Associations between polymorphisms in the myostatin, αA-globin and lactate dehydrogenase B genes and racing performance in homing pigeons. Vet. Med. (Praha) 2018, 63, 390–394. [Google Scholar] [CrossRef]
- Dybus, A.; Haase, E. Feather keratin gene polymorphism (f-ker) in domestic pigeons. Br. Poult. Sci. 2011, 52, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Proskura, W.S.; Kustosz, J.; Dybus, A.; Lanckriet, R. Polymorphism in dopamine receptor D4 gene is associated with pigeon racing performance. Anim. Genet. 2015, 46, 586–587. [Google Scholar] [CrossRef] [PubMed]
- Hubáček, J.A.; Pikhart, H.; Peasey, A.; Kubínová, R.; Bobák, M. Nobody is perfect: Comparison of the accuracy of PCR-RFLP and KASPTM method for genotyping. ADH1B and FTO polymorphisms as examples. Folia Biol. (Czech Republic) 2015, 61, 156–160. [Google Scholar] [PubMed]
- Mao, H.G.; Cao, H.Y.; Liu, H.H.; Dong, X.Y.; Xu, N.Y.; Yin, Z.Z. Association of ADSL gene polymorphisms with meat quality and carcass traits in domestic pigeons (Columba livia). Br. Poult. Sci. 2018, 59, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Chung, E.R. Association of SNP marker in the leptin gene with carcass and meat quality traits in Korean cattle. Asian-Australasian J. Anim. Sci. 2007, 20, 1–6. [Google Scholar] [CrossRef]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNPs and other types of molecular markers. Genet. Sel. Evol. 2008, 40, 241–264. [Google Scholar] [CrossRef]
- Pourzand, C.; Cerutti, P. Genotypic mutation analysis by RFLP/PCR. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1993, 288, 113–121. [Google Scholar] [CrossRef]
- Yang, C.H.; Cheng, Y.H.; Chuang, L.Y. A natural PCR-RFLP primer design for SNP genotyping using a genetic algorithm. In Proceedings of the International MultiConference of Engineers and Computer Scientists 2010, Hang Kong, China, 17–19 March 2010; pp. 124–129. [Google Scholar]
- Dybus, A.; Knapik, K. A new PCR-RFLP within the domestic pigeon (Columba livia var. domestica) cytochrome b (MTCYB) gene. J. Appl. Genet. 2005, 46, 315–317. [Google Scholar] [PubMed]
- Ota, M.; Fukushima, H.; Kulski, J.K.; Inoko, H. Single nucleotide polymorphism detection by polymerase chain reaction-restriction fragment length polymorphism. Nat. Protoc. 2007, 2, 2857–2864. [Google Scholar] [CrossRef]
- Kumpatla, S.P.; Buyyarapu, R.; Abdurakhmonov, I.Y.; Mammadov, J.A. Genomics-Assisted Plant Breeding in the 21st Century: Technological Advances and Progress. Plant Breed. 2010, 131–185. [Google Scholar] [CrossRef]
- Patterson, E.L.; Flemin, M.B.; Kessler, K.C.; Nissen, S.J.; Gaines, T.A. A KASP genotyping method to identify northern watermilfoil, Eurasian watermilfoil, and their interspecific hybrids. Front. Plant Sci. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Wosula, E.N.; Chen, W.; Amour, M.; Fei, Z.; Legg, J.P. KASP genotyping as a molecular tool for diagnosis of cassava-colonizing Bemisia tabaci. Insects 2020, 11, 305. [Google Scholar] [CrossRef]
- Liu, L.L.; Fang, C.; Ma, H.Y.; Yu, X.; Lv, S.P.; Liu, W.J. Development and validation of KASP markers for the milk traits genes in Kazakh horse. J. Appl. Anim. Res. 2020, 48, 293–299. [Google Scholar] [CrossRef]
- Tan, C.T.; Assanga, S.; Zhang, G.; Rudd, J.C.; Haley, S.D.; Xue, Q.; Ibrahim, A.; Bai, G.; Zhang, X.; Byrne, P.; et al. Development and validation of kasp markers for wheat streak mosaic virus resistance gene wsm2. Crop Sci. 2017, 57, 340–349. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, D.; Huang, H.; Yang, Z.; Wang, Y.; Yu, Y.; Liu, L.; Zhang, S.; Han, J.; Xiao, W. Technical note: Development and application of KASP assays for rapid screening of 8 genetic defects in Holstein cattle. J. Dairy Sci. 2020, 103, 619–624. [Google Scholar] [CrossRef]
- Zhao, S.; Li, A.; Li, C.; Xia, H.; Zhao, C.; Zhang, Y.; Hou, L.; Wang, X. Development and application of KASP marker for high throughput detection of AhFAD2 mutation in peanut. Electron. J. Biotechnol. 2017, 25, 9–12. [Google Scholar] [CrossRef]
- Cheon, K.S.; Baek, J.; Cho, Y.I.; Jeong, Y.M.; Lee, Y.Y.; Oh, J.; Won, Y.J.; Kang, D.Y.; Oh, H.; Kim, S.L.; et al. Single Nucleotide Polymorphism (SNP) discovery and Kompetitive Allele-Specific PCR (KASP) marker development with Korean Japonica rice varieties. Plant Breed. Biotechnol. 2018, 6, 391–403. [Google Scholar] [CrossRef]
- Corominas, J.; Ramayo-Caldas, Y.; Puig-Oliveras, A.; Pérez-Montarelo, D.; Noguera, J.L.; Folch, J.M.; Ballester, M. Polymorphism in the ELOVL6 Gene Is Associated with a Major QTL Effect on Fatty Acid Composition in Pigs. PLoS ONE 2013, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Devran, Z.; Göknur, A. Development and validation of a SNP-based KASP assay for rapid identification of Aphelenchoides besseyi Christie, 1942. Crop Prot. 2020, 136. [Google Scholar] [CrossRef]
- Graves, H.; Rayburn, A.L.; Gonzalez-Hernandez, J.L.; Gyoungju, N.; Do-Soon, K.; Lee, D.K. Validating DNA polymorphisms using KASP assay in prairie cordgrass (Spartina pectinata link) populations in the U.S. Front. Plant Sci. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.; Hubbart-Edwards, S.; Yang, C.; Devi, U.; Baker, L.; Heath, J.; Ashling, S.; Scholefield, D.; Howells, C.; Yarde, J.; et al. Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnol. J. 2020, 18, 743–755. [Google Scholar] [CrossRef]
- Kusza, S.; Cziszter, L.T.; Ilie, D.E.; Sauer, M.; Padeanu, I.; Gavojdian, D. Kompetitive Allele Specific PCR (KASPTM) genotyping of 48 polymorphisms at different caprine loci in French Alpine and Saanen goat breeds and their association with milk composition. PeerJ 2018, 2018. [Google Scholar] [CrossRef]
- Jedrzejczak-Silicka, M.; Lepczynski, A.; Gołębiowski, F.; Dolata, D.; Dybus, A. Application of PCR-HRM method for microsatellite polymorphism genotyping in the LDHA gene of pigeons (Columba livia). PLoS ONE 2021, 16, e0256065. [Google Scholar] [CrossRef] [PubMed]



| Gene | Sequence | Restriction Enzyme | Reference |
|---|---|---|---|
| LDHA | F 5′-TGAAGGGGTACACATCATGG-3′ R 5′-CCTTCTGGATTCCCCAGAGT-3′ | HaeIII | [9] |
| MTCYB | F 5′-TTTGGGTCCCTACTAGGCATT-3′ R 5′-GAGGACAAGGAGGATGGTGA-3′ | MvaI | [26] |
| DRD4 (1) | F 5′-TTTGGGATCGCTCGCTTACC-3′ R 5′-ATGACAGGGGATGCTACAGC-3′ | HpyCH4III | [19] |
| DRD4 (2) | F 5′-GGGCCAACAGGAAGCTCTAT-3′ R 5′-GCAGGACAACACAGCGTCTC-3′ | MnlI | [19] |
| Gene | KASP Assay | Position | Sequences (5′–3′) | Gene ID |
|---|---|---|---|---|
| LDHA | Allele-G Forward Primer | 52–76 | ATCTCTACAGTTGTTAAGGTGAGCG | MW072294.1 |
| Allele-A Forward Primer | 51–76 | AATCTCTACAGTTGTTAAGGTGAGCA | ||
| Common Reverse Primer | 122–94 | CCAAGGTTTTTAGGTCTCAGTAAGACAAA | ||
| MTCYB | Allele-C Forward Primer | 14314–14336 | ACTTCTCCCTAAAAGACATCCTC | NC013978.1 |
| Allele-G Forward Primer | 14312–14336 | CTACTTCTCCCTAAAAGACATCCTG | ||
| Common Reverse Primer | 14371–14347 | AGGGTCATTAGGGGGAGGAGTATTA | ||
| DRD4 (1) | Allele-T Reverse Primer | 45–26 | GAGCCAGGCCCAGGGTACTA | MT982613.1 |
| Allele-C Reverse Primer | 44–26 | AGCCAGGCCCAGGGTACTG | ||
| Common Forward Primer | 2–25 | CGCTTACCTTACGAGCGGTGACAA | ||
| DRD4 (2) | Allele-C Forward Primer | 524–504 | CGACTGTCTCCTATCCCCACC | MT982613.1 |
| Allele-T Forward Primer | 524–504 | CGACTGTCTCCTATCCCCACT | ||
| Common Reverse Primer | 575–554 | GGCCGTTGATCTTGGCCCGTTT |
| Gene | Genotype | PCR-RFLP | KASP | Percent Similarity |
|---|---|---|---|---|
| LDHA | AA | 1 | 1 | 100% |
| AG | 26 | 26 | 100% | |
| GG | 80 | 80 | 100% | |
| MTCYB | G | 1 | 1 | 100% |
| C | 106 | 106 | 100% | |
| DRD4 (1) | CC | 77 | 77 | 100% |
| CT | 26 | 26 | 100% | |
| TT | 4 | 4 | 100% | |
| DRD4 (2) | TT | 1 | 1 | 100% |
| TC | 12 | 12 | 100% | |
| CC | 94 | 94 | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-C.; Silva, B.B.I.; Huang, H.-Y.; Tsai, C.-Y.; Flores, R.J.D.; Tayo, L.L.; Tyan, Y.-C.; Tsai, M.-A.; Catulin, G.E.M.; Chuang, K.-P.; et al. Development and Validation of KASP Assays for the Genotyping of Racing Performance-Associated Single Nucleotide Polymorphisms in Pigeons. Genes 2021, 12, 1383. https://doi.org/10.3390/genes12091383
Chang C-C, Silva BBI, Huang H-Y, Tsai C-Y, Flores RJD, Tayo LL, Tyan Y-C, Tsai M-A, Catulin GEM, Chuang K-P, et al. Development and Validation of KASP Assays for the Genotyping of Racing Performance-Associated Single Nucleotide Polymorphisms in Pigeons. Genes. 2021; 12(9):1383. https://doi.org/10.3390/genes12091383
Chicago/Turabian StyleChang, Ching-Chi, Benji Brayan I. Silva, Huai-Ying Huang, Ching-Yi Tsai, Ronilo Jose D. Flores, Lemmuel L. Tayo, Yu-Chang Tyan, Ming-An Tsai, Gail Everette M. Catulin, Kuo-Pin Chuang, and et al. 2021. "Development and Validation of KASP Assays for the Genotyping of Racing Performance-Associated Single Nucleotide Polymorphisms in Pigeons" Genes 12, no. 9: 1383. https://doi.org/10.3390/genes12091383
APA StyleChang, C.-C., Silva, B. B. I., Huang, H.-Y., Tsai, C.-Y., Flores, R. J. D., Tayo, L. L., Tyan, Y.-C., Tsai, M.-A., Catulin, G. E. M., Chuang, K.-P., & Yang, J.-L. (2021). Development and Validation of KASP Assays for the Genotyping of Racing Performance-Associated Single Nucleotide Polymorphisms in Pigeons. Genes, 12(9), 1383. https://doi.org/10.3390/genes12091383









