Genome-Wide Identification and Expression Profiling of 2OGD Superfamily Genes from Three Brassica Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome-Wide Investigation of 2OGD Genes in Three Brassica Plants
2.2. Sequences and Chromosomal Location Analyses
2.3. Gene Structural Determination and Subcellular Location
2.4. Sequence Homologous and Phylogenetic Analyses of 2OGD
2.5. Gene Expression Analysis in Different Tissues
2.6. Expression Analysis by qRT-PCR
3. Results
3.1. Genome-Wide Identification of 2OGD Members in Brassica Plants
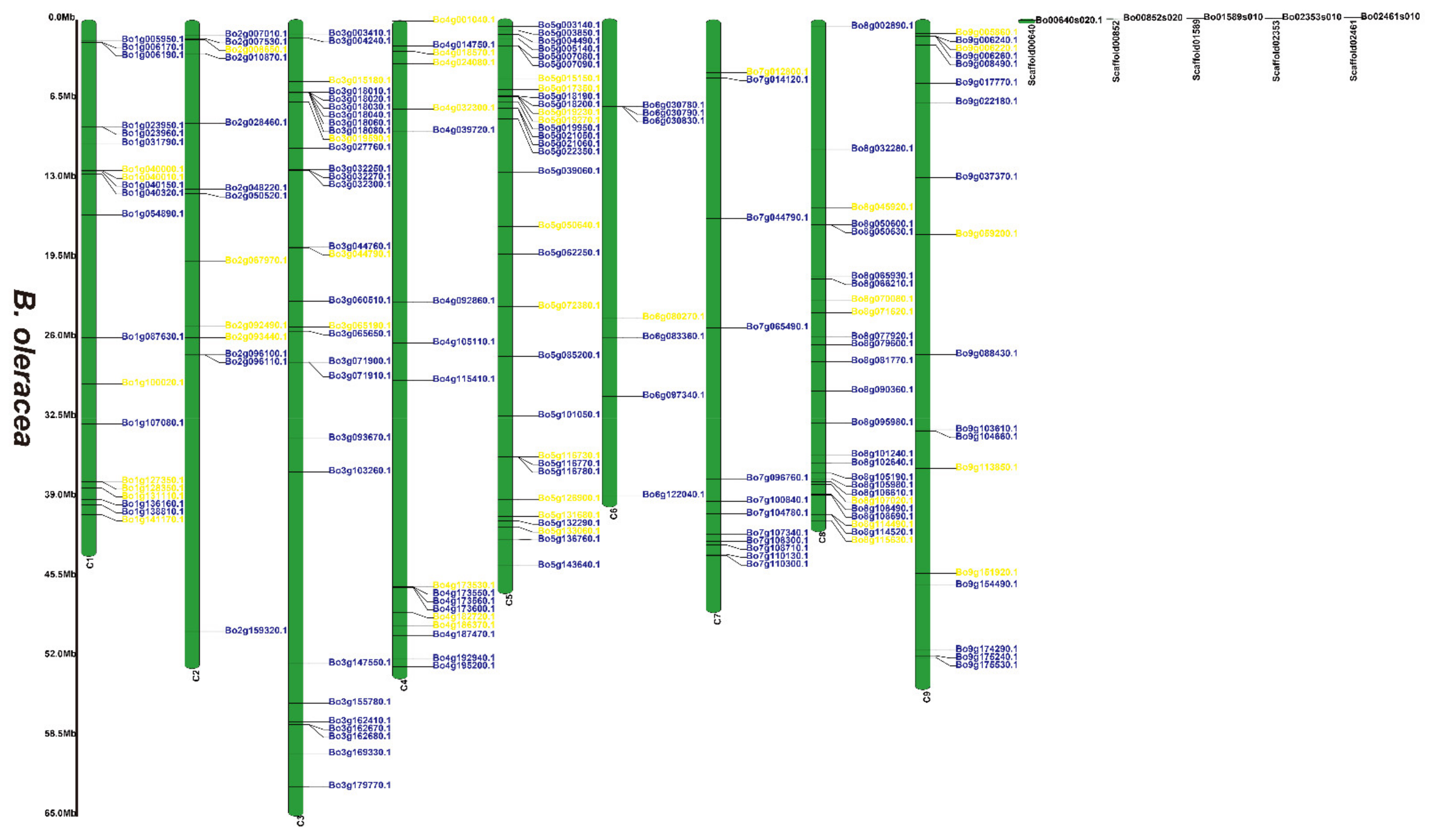
3.2. Chromosomal Locations of the 2OGD Gene Family
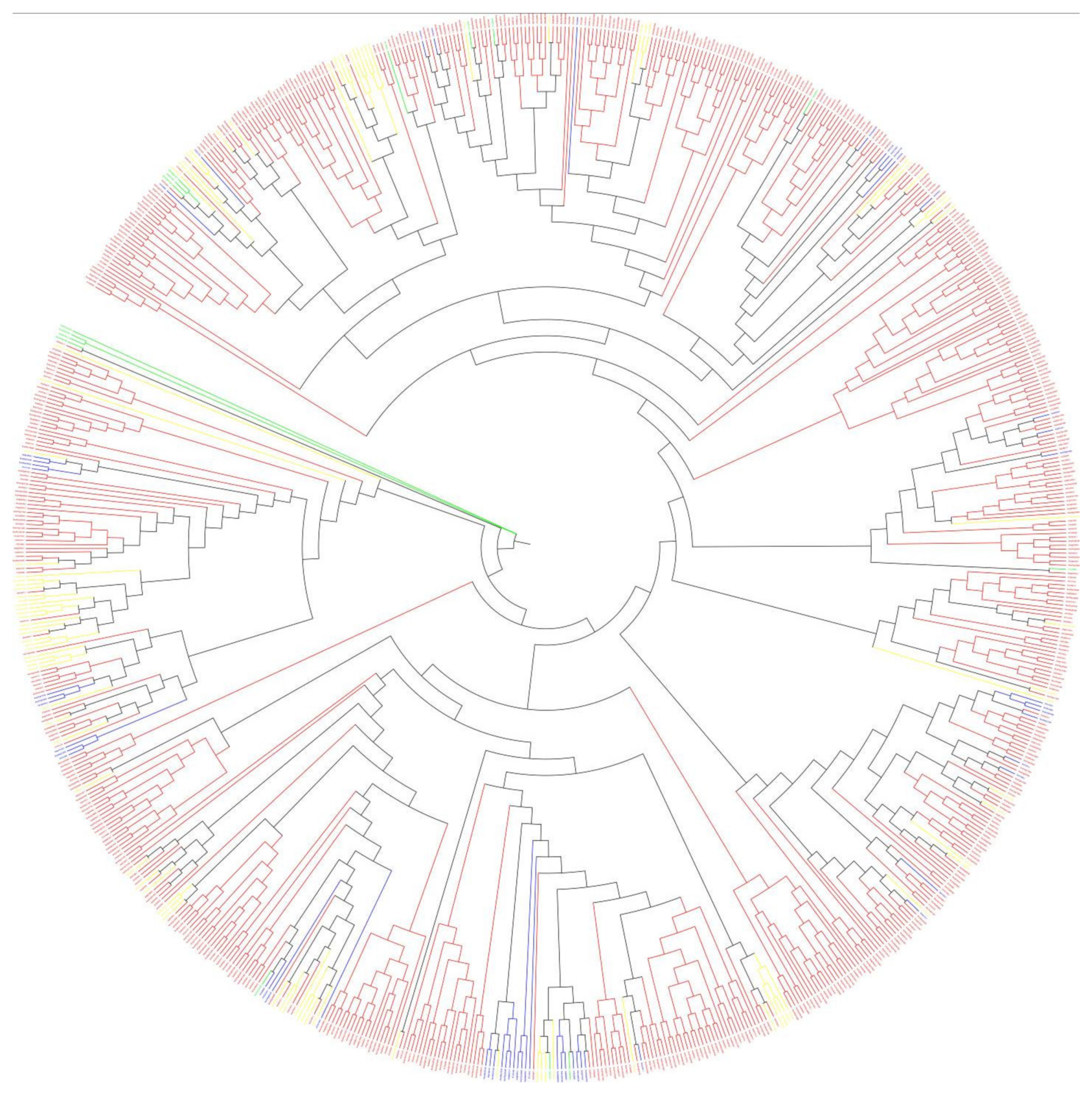
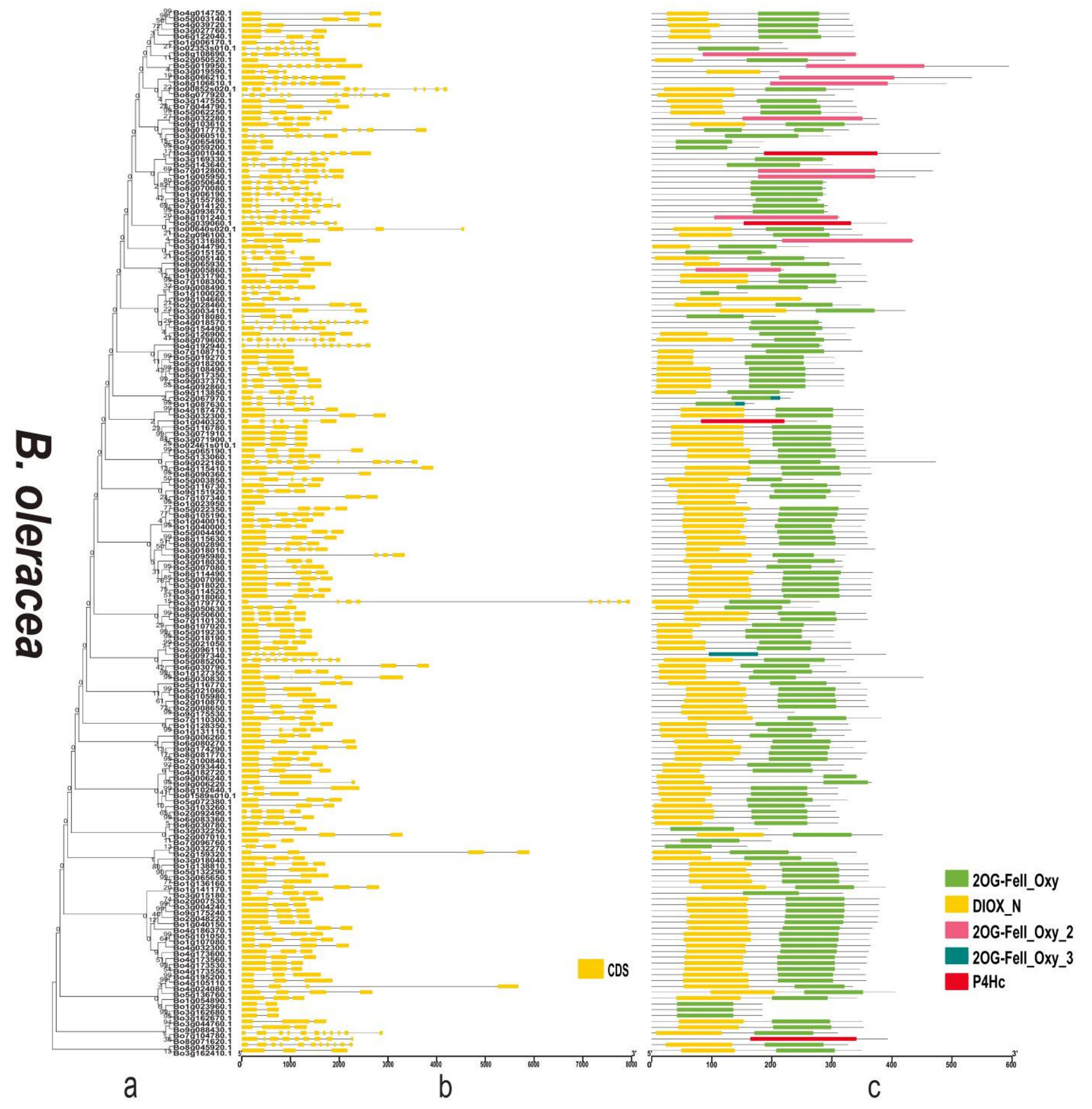
3.3. Phylogenetic Analysis and Diversity of 2OGDs
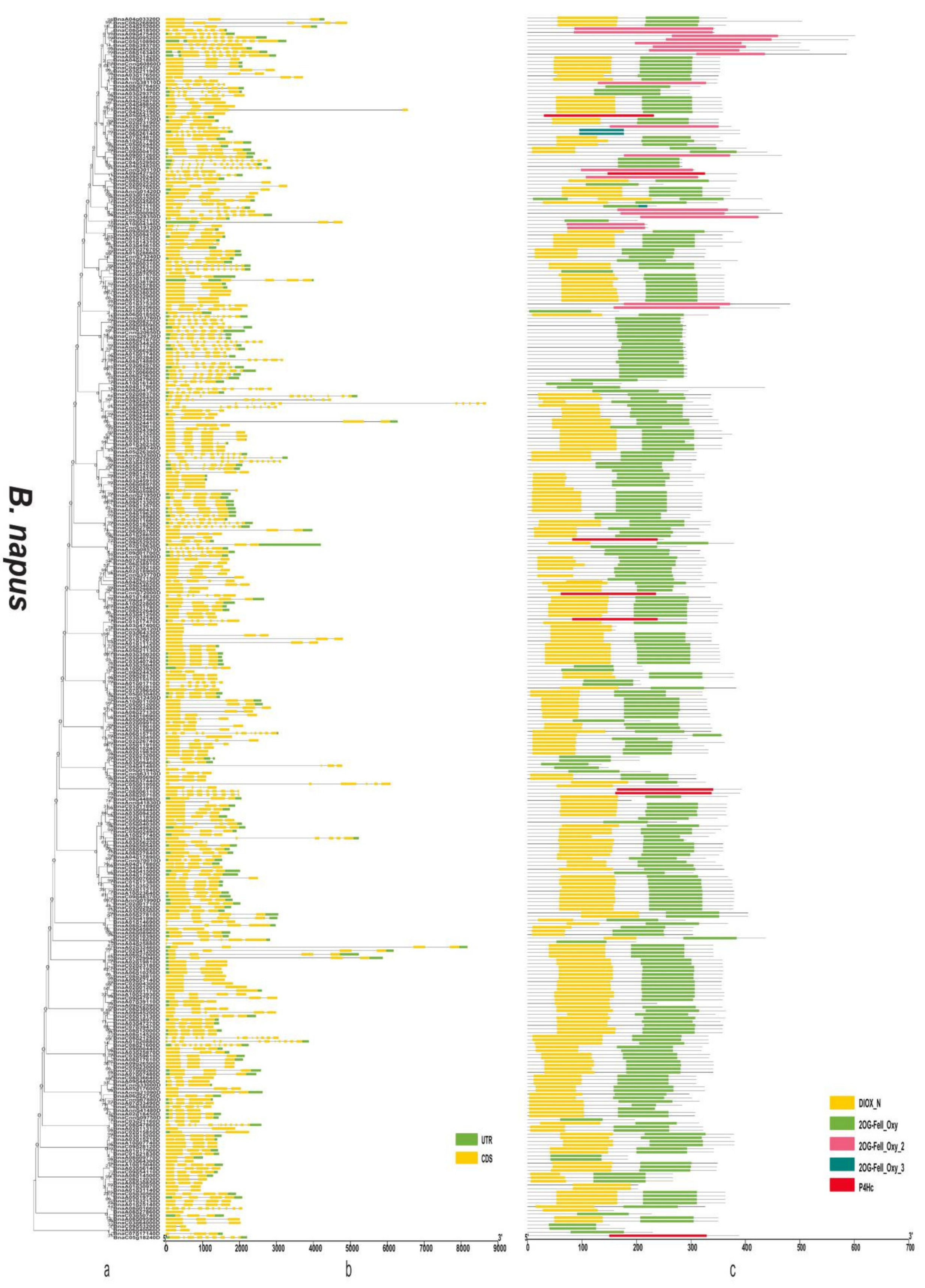
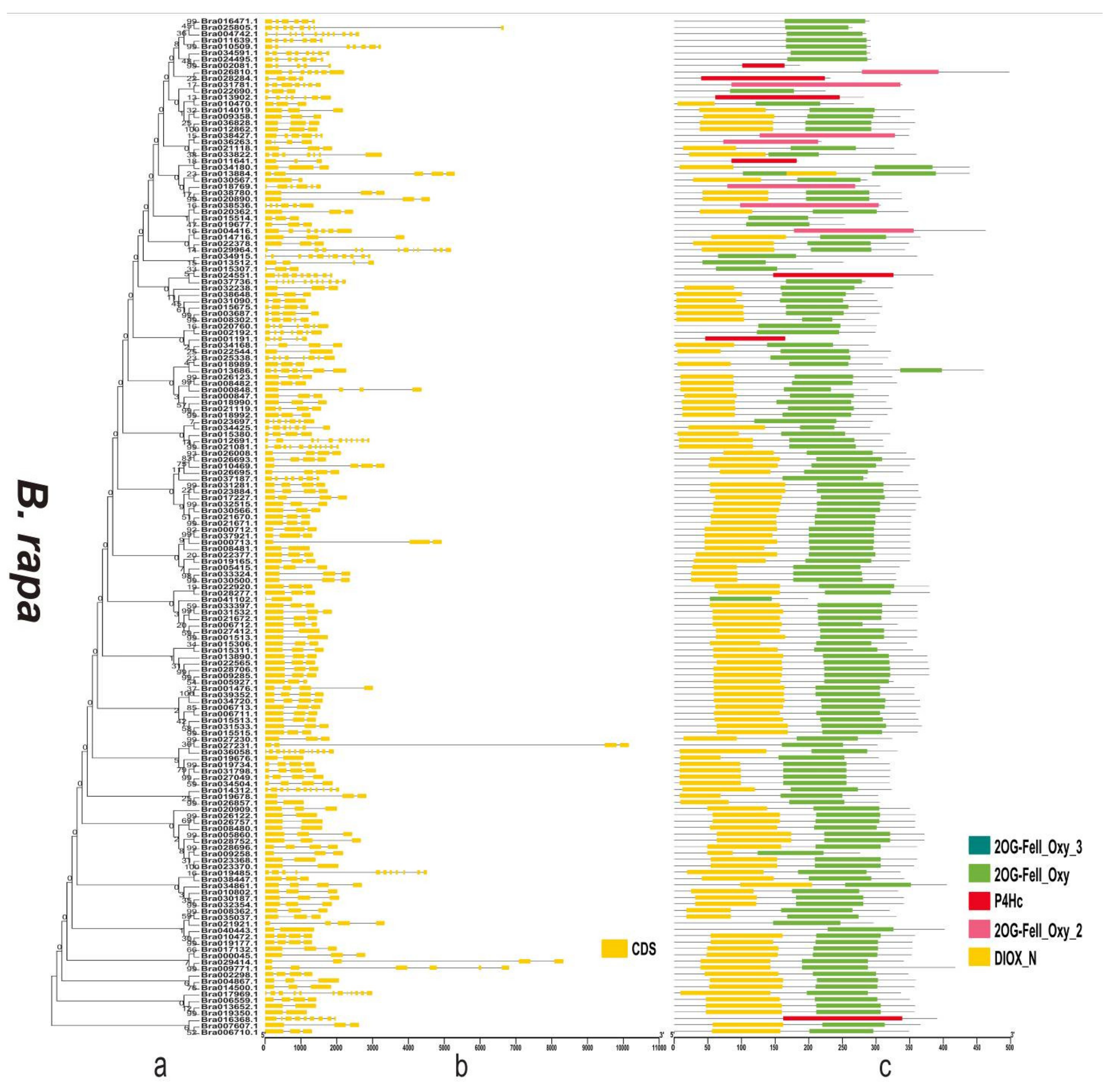
3.4. Gene Structure and Domain Analysis of the 2OGD Gene Family
3.5. Differentially Expressed A. thaliana 2OGD Genes at Different Developmental Stages
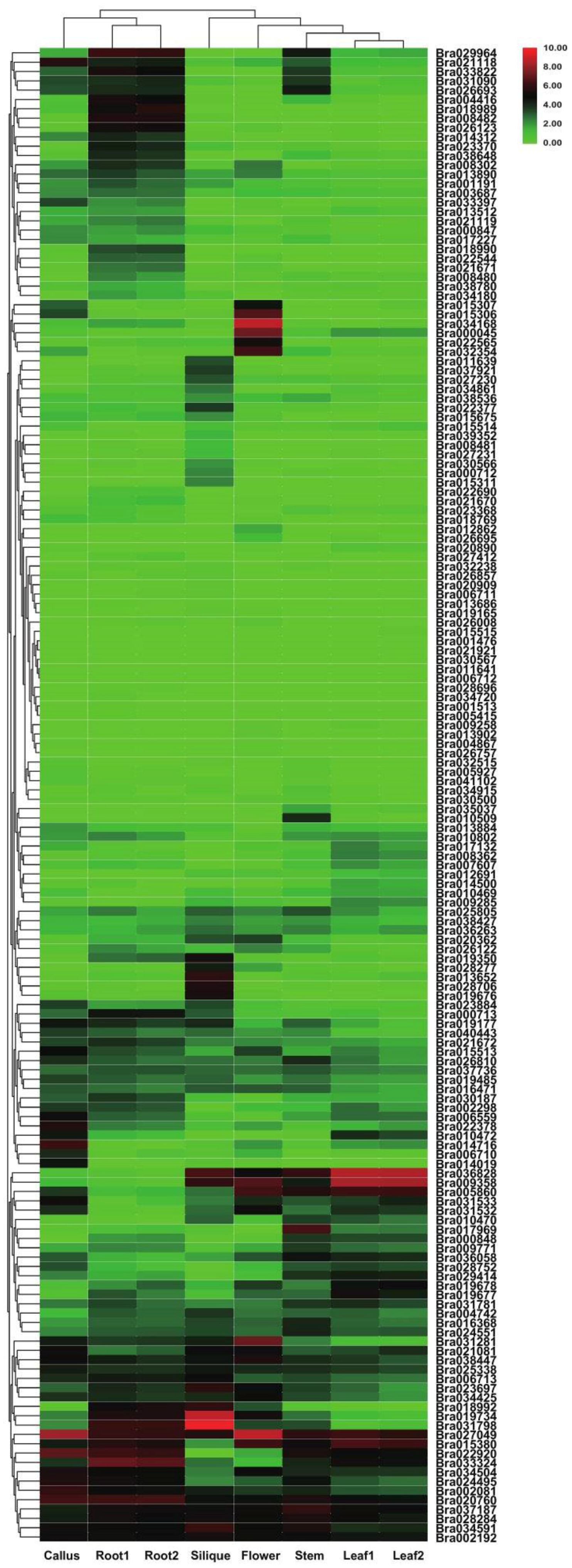
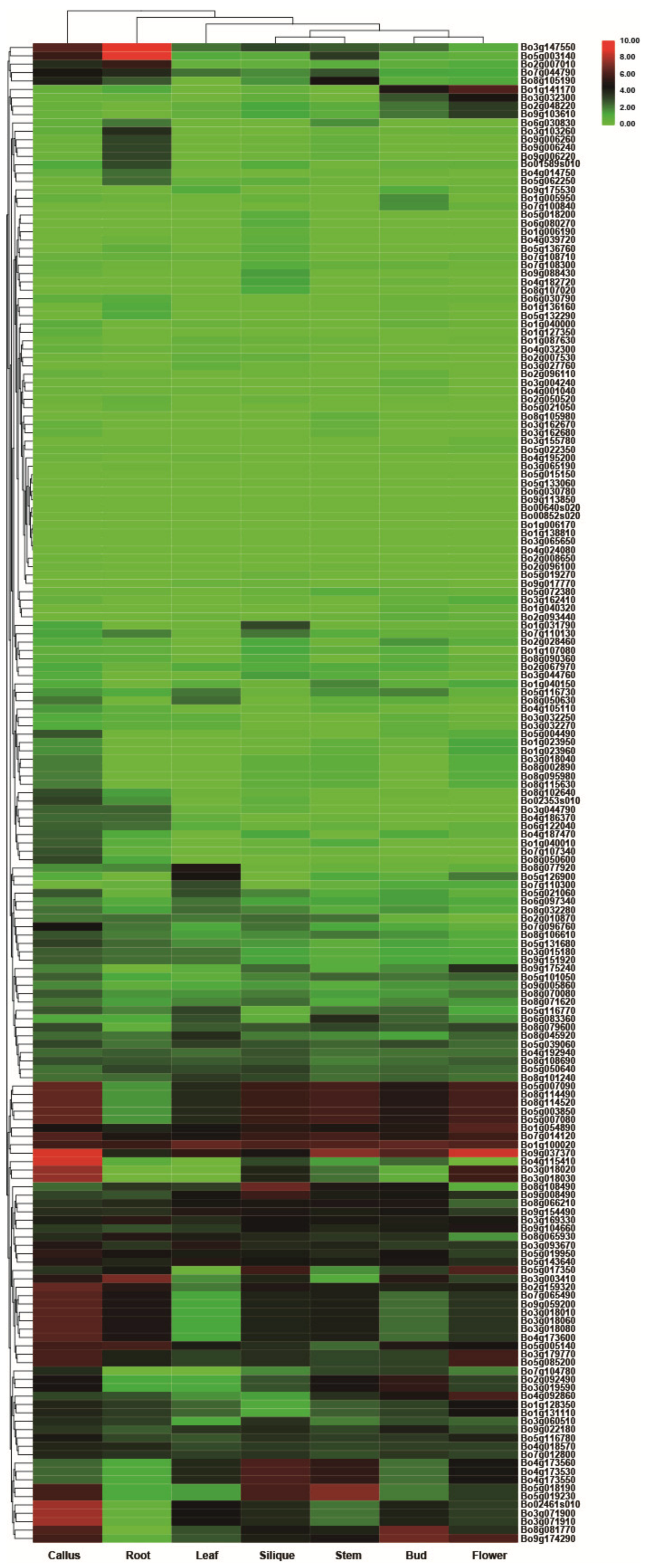
3.6. Transcriptome Analysis of Tissue-Specific Expression
3.7. qRT-PCR Analysis in Different Tissues
4. Discussion
4.1. Evolution and Expansion of 2OGD Genes
4.2. Gene Expression and Transcriptome Analysis Could Better Predict Potential Gene Functions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Hangasky, J.A.; Taabazuing, C.Y.; Valliere, M.A.; Knapp, M.J. Imposing function down a (cupin)-barrel: Secondary structure and metal stereochemistry in the αKG-dependent oxygenases. Metallomics 2013, 5, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Flashman, E.; Schofield, C.J. The most versatile of all reactive intermediates? Nat. Chem. Biol. 2007, 3, 86–87. [Google Scholar] [CrossRef]
- Zhang, Z.; Ren, J.S.; Clifton, I.J.; Schofield, C.J. Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase-the ethylene-forming enzyme. Chem. Biol. 2004, 11, 1383–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.D.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 2002, 10, 93–103. [Google Scholar] [CrossRef]
- Hedden, P. 2-Oxoglutarate-dependent dioxygenases in plants: Mechanism and function. Biochem. Soc. Trans. 1992, 20, 373–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagel, J.M.; Facchini, P.J. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 2010, 6, 273–275. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001, 2, research0007.1. [Google Scholar] [CrossRef]
- De Carolis, E.; De Luca, V. 2-Oxoglutarate-dependent dioxygenase and related enzymes: Biochemical characterization. Phytochemistry 1994, 36, 1093–1107. [Google Scholar] [CrossRef]
- Falnes, P.Ø.; Johansen, R.F.; Seeberg, E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002, 419, 178–182. [Google Scholar] [CrossRef]
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002, 419, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Keskiaho, K.; Hieta, R.; Sormunen, R.; Myllyharju, J. Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 2007, 19, 256–269. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, A.J.; Bouzayen, M.; Grierson, D. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc. Natl. Acad. Sci. USA 1991, 88, 7434–7437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbull, J.J.; Nakajima, J.-I.; Welford, R.W.D.; Yamazaki, M.; Saito, K.; Schofield, C.J. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: Anthocyanidin synthase, flavonol synthase, and flavanone 3beta–hydroxylase. J. Biol. Chem. 2004, 279, 1206–1216. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Kobayashi, M.; Gong, Z.; Tanaka, Y.; Yamazaki, M. Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: Molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 1999, 17, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Holton, T.A.; Brugliera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Shen, G.; Pang, Y.; Wu, W.; Deng, Z.; Zhao, L.; Cao, Y.; Sun, X.; Tang, K. Cloning and characterization of a flavanone 3-hydroxylase gene from Ginkgo biloba. Biosci. Rep. 2006, 26, 19–29. [Google Scholar] [CrossRef]
- Jackson, R.G.; Lim, E.K.; Li, Y.; Kowalczyk, M.; Sandberg, G.; Hoggett, J.; Ashford, D.A.; Bowles, D.J. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 2001, 276, 4350–4356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in Rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giamoustaris, A.; Mithen, R. The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica napus ssp. oleifera) on its interaction with specialist and generalist pests. Ann. Appl. Biol. 1995, 126, 347–363. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Lambrix, V.M.; Reichelt, M.; Gershenzon, J.; MitchellOlds, T. Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 2001, 13, 681–693. [Google Scholar] [CrossRef] [Green Version]
- Hansen, B.G.; Kerwin, R.E.; Ober, J.A.; Lambrix, V.M.; Mitchell-Olds, T.; Gershenzon, J.; Halkier, B.A.; Kliebenstein, D.J. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008, 148, 2096–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakizaki, T.; Kitashiba, H.; Zou, Z.; Li, F.; Fukino, N.; Ohara, T.; Nishio, T.; Ishida, M. A 2-oxoglutarate-dependent dioxygenase mediates the biosynthesis of glucoraphasatin in Radish. Plant Physiol. 2017, 173, 1583–1593. [Google Scholar] [CrossRef] [Green Version]
- Farrow, S.C.; Facchini, P.J. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant. Sci. 2014, 5, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2007, 36, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2006, 35, 883–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Song, J. The 2-oxoglutarate-dependent dioxygenase superfamily participates in tanshinone production in Salvia miltiorrhiza. J. Exp. Bot. 2017, 68, 2299–2308. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB transcription factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Lei, J.; Cao, B.; Wu, S.; Chen, G.; Chen, C. Molecular cloning and characterization of three glucosinolate transporter (GTR) genes from Chinese kale. Genes 2019, 10, 202. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Chen, G.; Chen, C.; Cao, B. Germplasm diversity of Chinese kale in China. Hortic. Plant J. 2017, 3, 101–104. [Google Scholar] [CrossRef]
- Wu, S.; Lei, J.; Chen, G.; Chen, H.; Cao, B.; Chen, C. De novo transcriptome assembly of Chinese kale and global expression analysis of genes involved in glucosinolate metabolism in multiple tissues. Front. Plant Sci. 2017, 8, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schwechheimer, C. Understanding gibberellic acid signaling—are we there yet? Curr. Opin Plant. Biol. 2008, 11, 9–15. [Google Scholar] [CrossRef]
- Yang, T.J.; Kim, J.S.; Kwon, S.J.; Lim, K.B.; Cell, B.-S.P.J.P. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 2006, 18, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Shao, Z.Q.; Wu, X.Z.; Wang, Q.; Wang, B.; Chen, J.Q.; Hang, Y.Y.; Xue, J.Y. Loss/retention and evolution of NBS-encoding genes upon whole genome triplication of Brassica rapa. Gene 2014, 540, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lyu, T.; Xu, L.; Hu, Z.; Xiong, X.; Liu, T.; Cao, J. Complex molecular evolution and expression of expansin gene families in three basic diploid species of Brassica. Int. J. Mol. Sci. 2020, 21, 3424. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.; Qin, R.; Hua, W.; et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014, 15, 3. [Google Scholar] [CrossRef] [Green Version]
- Papp, B.; Pal, C.; Hurst, L.D. Dosage sensitivity and the evolution of gene families in yeast. Nature 2003, 424, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.A.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Xie, F.; Zhang, Y.; Tang, H. Identification and evolution of GRAS transcription factor family in four Brassica species. AIP Conf. Proc. 2019, 2058, 020028. [Google Scholar] [CrossRef]
- Lohani, N.; Golicz, A.A.; Singh, M.B.; Bhalla, P.L. Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus. Funct. Integr. Genom. 2019, 19, 515–531. [Google Scholar] [CrossRef]
- Kataoka, H.; Yamamoto, Y.; Sekiguchi, M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 1983, 153, 1301–1307. [Google Scholar] [CrossRef] [Green Version]
- Ozer, A.; Bruick, R.K. Non-heme dioxygenases: Cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 2007, 3, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Hieta, R.; Myllyharju, J. Cloning and characterization of a low molecular weight prolyl 4–hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J. Biol. Chem. 2002, 277, 23965–23971. [Google Scholar] [CrossRef] [Green Version]
- Hudgins, J.W.; Ralph, S.G.; Franceschi, V.R.; Bohlmann, J. Ethylene in induced conifer defense: cDNA cloning, protein expression, and cellular and subcellular localization of 1-aminocyclopropane-1-carboxylate oxidase in resin duct and phenolic parenchyma cells. Planta 2006, 224, 865–877. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, A.L.; Ward, D.A.; Uknes, S.; Appleford, N.E.; Lange, T.; Huttly, A.K.; Gaskin, P.; Graebe, J.E.; Hedden, P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995, 108, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Hedden, P.; Phillips, A.L.; Rojas, M.C.; Carrera, E.; Tudzynski, B. Gibberellin biosynthesis in plants and fungi: A case of convergent evolution? J. Plant Growth Regul. 2002, 20, 319–331. [Google Scholar] [CrossRef]
- Chiang, H.H.; Hwang, I.; Goodman, H.M. Isolation of the Arabidopsis GA4 locus. Plant Cell 1995, 7, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.; Phillips, A.L.; Gaskin, P.; Hedden, P. Function and substrate specificity of the gibberellin 3beta-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol. 1998, 117, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Li, G.; Chen, G.; Lei, J.; Cao, B.; Chen, C. Genome-Wide Identification and Expression Profiling of 2OGD Superfamily Genes from Three Brassica Plants. Genes 2021, 12, 1399. https://doi.org/10.3390/genes12091399
Jiang D, Li G, Chen G, Lei J, Cao B, Chen C. Genome-Wide Identification and Expression Profiling of 2OGD Superfamily Genes from Three Brassica Plants. Genes. 2021; 12(9):1399. https://doi.org/10.3390/genes12091399
Chicago/Turabian StyleJiang, Ding, Guangguang Li, Guoju Chen, Jianjun Lei, Bihao Cao, and Changming Chen. 2021. "Genome-Wide Identification and Expression Profiling of 2OGD Superfamily Genes from Three Brassica Plants" Genes 12, no. 9: 1399. https://doi.org/10.3390/genes12091399
APA StyleJiang, D., Li, G., Chen, G., Lei, J., Cao, B., & Chen, C. (2021). Genome-Wide Identification and Expression Profiling of 2OGD Superfamily Genes from Three Brassica Plants. Genes, 12(9), 1399. https://doi.org/10.3390/genes12091399





