Influence of DAT1 Promotor Methylation on Sports Performance
Abstract
:1. Introduction
1.1. DNA Methylation
1.2. DAT1
2. Materials and Methods
2.1. Samples
2.2. Methylation Status Assessment
2.3. Assessment of the Ability to Bind Transcription Factors
2.4. Psychometric Tests
2.5. Statistical Analysis
3. Results
3.1. PAX 5 CpG Position: Sites 3
3.2. PAX 5 CpG Position: Sites 13
3.3. PAX 5 CpG Position: Sites 22
3.4. Neuroticism Scale
3.5. Extraversion Scale
3.6. Openness Scale
3.7. Agreeability Scale
3.8. PAX 5 CpG Position: Sites 33
4. Discussion
DAT1 and Methylation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2012, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.-J.; Chang, L.; Volkow, N.D.; Telang, F.; Logan, J.; Ernst, T.; Fowler, J.S. Decreased brain dopaminergic transporters in HIV-associated dementia patients. Brain 2004, 127, 2452–2458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deaton, A.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levenson, J.M.; Roth, T.L.; Lubin, F.D.; Miller, C.; Huang, I.-C.; Desai, P.; Malone, L.M.; Sweatt, J.D. Evidence That DNA (Cytosine-5) Methyltransferase Regulates Synaptic Plasticity in the Hippocampus. J. Biol. Chem. 2006, 281, 15763–15773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, E.D.; Kavalali, E.T.; Monteggia, L.M. Activity-Dependent Suppression of Miniature Neurotransmission through the Regulation of DNA Methylation. J. Neurosci. 2008, 28, 395–406. [Google Scholar] [CrossRef] [Green Version]
- Frazer, K.A.; Ballinger, D.G.; Cox, D.R.; Hinds, D.A.; Stuve, L.L.; Gibbs, R.A.; Belmont, J.W.; Boudreau, A.; Hardenbol, P.; Leal, S.M.; et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851–861. [Google Scholar] [PubMed]
- Miller, C.; Campbell, S.L.; Sweatt, J.D. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn. Mem. 2008, 89, 599–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGowan, P.O.; Meaney, M.J.; Szyf, M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Res. 2008, 1237, 12–24. [Google Scholar] [CrossRef] [Green Version]
- Cirulli, F.; Francia, N.; Berry, A.; Aloe, L.; Alleva, E.; Suomi, S.J. Early life stress as a risk factor for mental health: Role of neuro-trophins from rodents to non-human primates. Neurosci. Biobehav. Rev. 2009, 33, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Barrès, R.; Zierath, J.R. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat. Rev. Endocrinol. 2016, 12, 441–451. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.L.; Walder, K.R. Exercise and the Skeletal Muscle Epigenome. Cold Spring Harb. Perspect. Med. 2017, 7, a029876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehler-Wex, C.; Riederer, P.; Gerlach, M. Dopaminergic dysbalance in distinct basal ganglia neurocircuits: Implications for the pathophysiology of Parkinson’s disease, schizophrenia and attention deficit hyperactivity disorder. Neurotox. Res. 2006, 10, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, T.A.; Schork, N.J.; Eskin, E.; Kelsoe, J.R. Identification of additional variants within the human dopamine transporter gene provides further evidence for an association with bipolar disorder in two independent samples. Mol. Psychiatry 2006, 11, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michałowska-Sawczyn, M.; Grzywacz, A.; Masiak, J.; Chmielowiec, K.; Chmielowiec, J.; Chycki, J.; Maculewicz, E.; Cięszczyk, P. Associations Between Physical Effort and DNA Methylation in the Promotor Region of the Dopamine Transporter Gene (DAT1). J. Hum. Kinet. 2021, 77, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; McCrae, R.R. The Revised NEO Personality Inventory (NEO-PI-R); Sage Pub-lications Inc.: Thousand Oaks, CA, USA, 2008; Volume 2, pp. 179–198. [Google Scholar]
- Drgon, T.; Lin, Z.; Wang, G.-J.; Fowler, J.; Pablo, J.; Mash, D.C.; Volkow, N.; Uhl, G.R. Common Human 5′ Dopamine Transporter (SLC6A3) Haplotypes Yield Varying Expression Levels In Vivo. Cell. Mol. Neurobiol. 2006, 26, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Martinat, C.; Bacci, J.-J.; Leete, T.; Kim, J.; Vanti, W.B.; Newman, A.H.; Cha, J.H.; Gether, U.; Wang, H.; Abeliovich, A. Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proc. Natl. Acad. Sci. USA 2006, 103, 2874–2879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberini, C.M. Transcription Factors in Long-Term Memory and Synaptic Plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Shumay, E.; Fowler, J.S.; Volkow, N.D. Genomic Features of the Human Dopamine Transporter Gene and Its Potential Epi-genetic States: Implications for Phenotypic Diversity. PLoS ONE 2010, 5, e11067. [Google Scholar] [CrossRef] [Green Version]
- Dolfini, D.; Mantovani, R.; Zambelli, F.; Pavesi, G. A perspective of promoter architecture from the CCAAT box. Cell Cycle 2009, 8, 4127–4137. [Google Scholar] [CrossRef] [Green Version]
- Finotti, A.; Treves, S.; Zorzato, F.; Gambari, R.; Feriotto, G. Upstream stimulatory factors are involved in the P1 promoter directed transcription of the A beta H-J-J locus. BMC Mol. Biol. 2008, 9, 110. [Google Scholar] [CrossRef] [Green Version]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.; Taylor, M.; Engström, P.; Frith, M.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef]
- Schulz, R.; McCole, R.B.; Woodfine, K.; Wood, A.J.; Chahal, M.; Monk, D.; Moore, G.E.; Oakey, R.J. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum. Mol. Genet. 2008, 18, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q. Transcriptional regulation of neuronal phenotype in mammals. J. Physiol. 2006, 575, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.J.; Trinklein, N.D.; Anton, E.D.; Nguyen, L.; Myers, R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2005, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelko, I.N.; Mueller, M.R.; Folz, R.J. Transcription Factors Sp1 and Sp3 Regulate Expression of Human Extracellular Superoxide Dismutase in Lung Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2008, 39, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radwan, A.; Younis, A.; Luykx, P.; Khuri, S. Prediction and analysis of nucleosome exclusion regions in the human genome. BMC Genom. 2008, 9, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, K.; Yang, J.; Reynolds, G.P.; Chen, B.; Shao, J.; Liu, R.; Qian, Q.; Liu, H.; Yang, R.; Wen, J.; et al. DAT1 methylation is associated with methylphenidate response on oppositional and hyperactive-impulsive symptoms in children and ado-lescents with ADHD. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry 2017, 18, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Rajala, A.Z.; Zaitoun, I.; Henriques, J.B.; Converse, A.K.; Murali, D.; Epstein, M.L.; Populin, L.C. Dopamine transporter gene susceptibility to methylation is associated with impulsivity in nonhuman primates. J. Neurophysiol. 2014, 112, 2138–2146. [Google Scholar] [CrossRef] [Green Version]
- Eberhard, D.; Jiménez, G.; Heavey, B.; Busslinger, M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000, 19, 2292–2303. [Google Scholar] [CrossRef] [Green Version]
- Abdolmaleky, H.M.; Smith, C.L.; Zhou, J.-R.; Thiagalingam, S. Epigenetic Alterations of the Dopaminergic System in Major Psychiatric Disorders. Methods Mol. Biol. 2008, 448, 187–212. [Google Scholar] [CrossRef]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J. DNA Methylation of the First Exon Is Tightly Linked to Transcriptional Silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Ferris, M.J.; Espana, R.A.; Locke, J.L.; Konstantopoulos, J.K.; Rose, J.H.; Chen, R.; Jones, S.R. Dopamine transporters govern diurnal variation in extracellular dopamine tone. Proc. Natl. Acad. Sci. USA 2014, 111, E2751–E2759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkow, N.D.; Ding, Y.S.; Fowler, J.S.; Wang, G.J.; Logan, J.; Gatley, S.J.; Hitzemann, R.; Smith, G.; Fields, S.D.; Gur, R. Dopamine transporters decrease with age. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 1996, 37, 554–559. [Google Scholar]
- Newberg, A.; Lerman, C.; Wintering, N.; Ploessl, K.; Mozley, P.D. Dopamine transporter binding in smokers and non-smokers. Clin. Nucl. Med. 2007, 32, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Wang, G.-J.; Smith, L.; Fowler, J.S.; Telang, F.; Logan, J.; Tomasi, D. Recovery of dopamine transporters with methamphetamine detoxification is not linked to changes in dopamine release. NeuroImage 2015, 121, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Ashok, A.H.; Mizuno, Y.; Volkow, N.D.; Howes, O.D. Association of Stimulant Use with Dopaminergic Alterations in Users of Cocaine, Amphetamine, or Methamphetamine: A Systematic Review and Meta-analysis. JAMA Psychiatry 2017, 74, 511–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.-J.; Volkow, N.D.; Wigal, T.; Kollins, S.H.; Newcorn, J.H.; Telang, F.; Logan, J.; Jayne, M.; Wong, C.T.; Han, H.; et al. Long-Term Stimulant Treatment Affects Brain Dopamine Transporter Level in Patients with Attention Deficit Hyperactive Disorder. PLoS ONE 2013, 8, e63023. [Google Scholar] [CrossRef] [Green Version]

| CpG Site | Studied Group | Methylation Status (%) | χ2(p) | OR | 95% CI (−95%, +95%) |
|---|---|---|---|---|---|
| 3 * | sports subjects N (100) | 78% | 20.471 (0.00001) | 4.838 | (2.326; 10.065) |
| control N (239) | 94% | ||||
| 13 * | sports subjects N (100) | 28% | 37.290 (0.00001) | 0.126 | (0.059; 0.265) |
| control N (239) | 5% | ||||
| 22 | sports subjects N (100) | 95% | 0.001 (0.974) | 0.982 | (0.337; 2.865) |
| control N (239) | 95% | ||||
| 33 | sports subjects N (100) | 66% | 9.291 (0.0023) | 2.247 | (1.326; 3.810) |
| control N (239) | 81% |
| STAI/NEO Five Factor Inventory/ | Sports Subjects (N = 100) | Control (N = 236) | Z | p Value |
|---|---|---|---|---|
| Neuroticism/scale | 4.76 ± 2.28 | 4.66 ± 1.99 | 0.145 | 0.884 |
| Extraversion/scale | 6.27 ± 1.89 | 6.41 ± 1.97 | −0.692 | 0.488 |
| Openness/scale | 4.44 ± 1.63 | 4.83 ± 1.70 | −1.531 | 0.126 |
| Agreeability/scale | 5.23 ± 2.13 | 5.65 ± 2.07 | −1.428 | 0.153 |
| Conscientiousness/scale | 7.23 ± 1.86 | 5.83 ± 2.13 | 5.498 | 0.0000 * |
| STAI/NEO Five-Factor Inventory/ | ANOVA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sports (N = 100) M ± SD | Control (N = 236) M ± SD | Methylation Status No (N = 17) M ± SD | Methylation Status Yes (N = 319) M ± SD | Full Model F p Value R2 | Factor | F (p Value) | ɳ2 | Power (alfa = 0.05) | |
| PAX 5 CpG Position: sites 3 | |||||||||
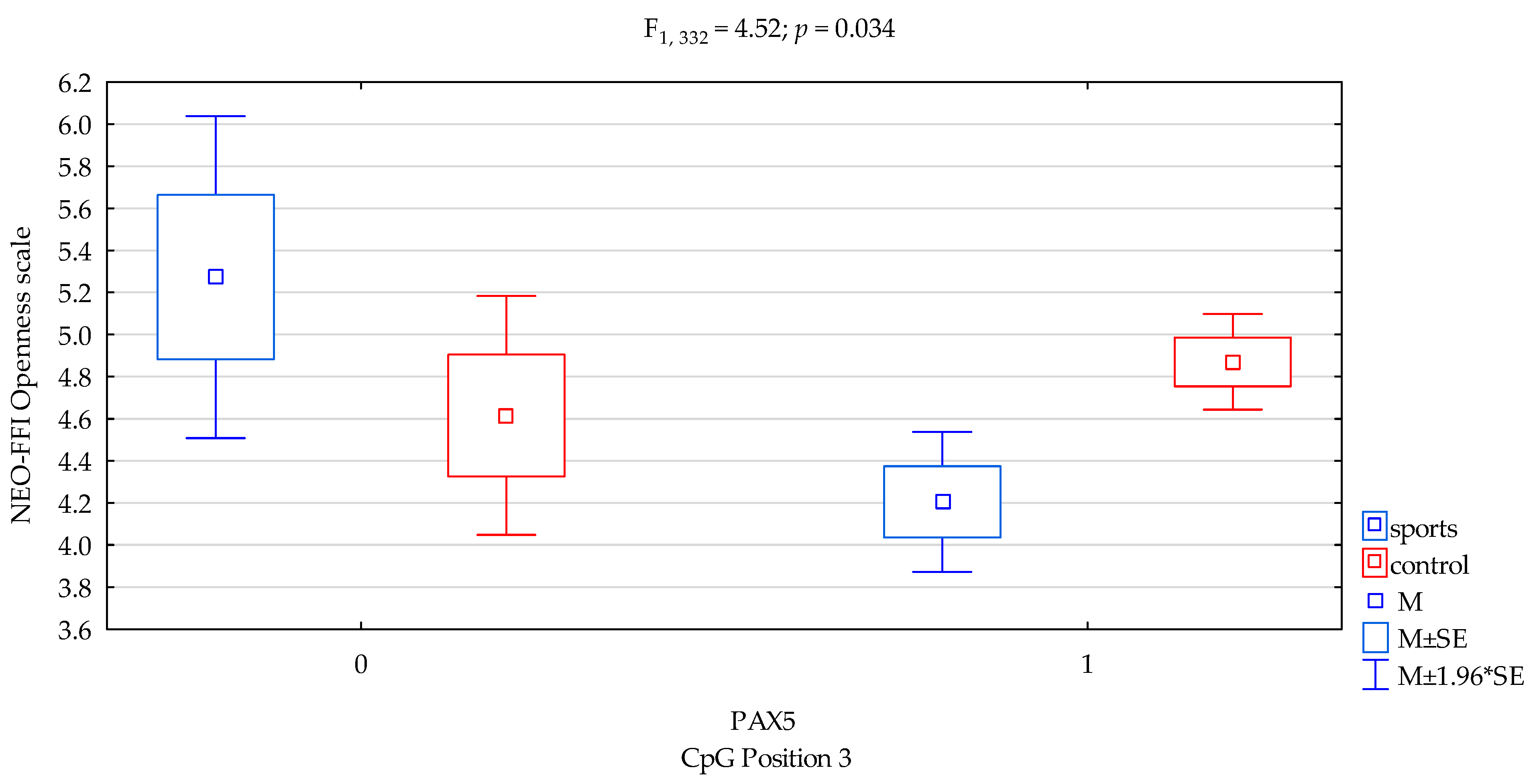
| Openness/scale | 4.44 ± 1.63 | 4.83 ± 1.70 | 5.02 ± 1.60 | 4.70 ± 1.69 | F3,332 = 3.913 p = 0.0091 * R2 = 0.034 | intercept | F1,332 = 929.65 (p < 0.0001) | 0.737 | 1.000 |
| sports/control | F1,332 = 0.0001 (p = 0.990) | 0.00001 | 0.050 | ||||||
| CpG sites 3 | F2,332 = 1.71 (p = 0.192) | 0.005 | 0.256 | ||||||
| addicts/control x CpG sites 3 | F2,332 = 4.52 (p = 0.034) * | 0.013 | 0.563 | ||||||
| PAX 5 CpG Position: sites 13 | |||||||||
| Neuroticism/scale | 4.76 ± 2.28 | 4.66 ± 1.99 | 4.69 ± 2.04 | 4.79 ± 2.34 | F3,332 = 2.687 p = 0.0465 * R2 = 0.024 | intercept | F1,332 = 526.07 (p < 0.0001) * | 0.613 | 1.000 |
| sports/control | F1,332 = 5.17 (p = 0.024) * | 0.015 | 0.621 | ||||||
| CpG sites 13 | F2,332 = 0.48 (p = 0.488) | 0.001 | 0.107 | ||||||
| addicts/control x CpG sites 13 | F2,332 = 7.89 (p = 0.005) * | 0.023 | 0.800 | ||||||
| PAX 5 CpG Position: sites 22 | |||||||||
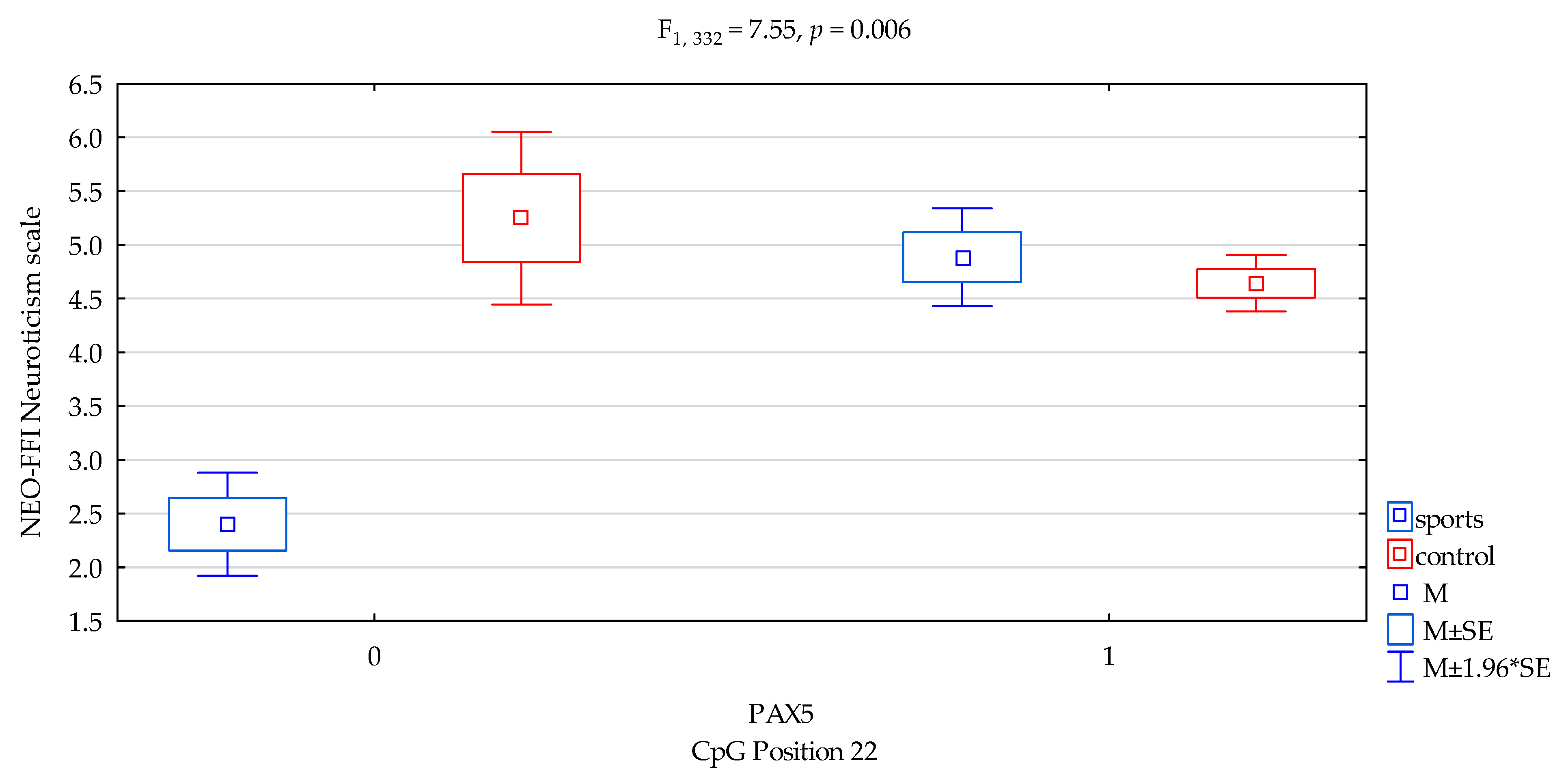
| Neuroticism/scale | 4.76 ± 2.28 | 4.66 ± 1.99 | 4.41 ± 1.80 | 4.71 ± 2.08 | F5,332 = 2.674 p = 0.0473 * R2 = 0.024 | intercept | F1,332 = 233.11 (p < 0.0001) * | 0.412 | 1.000 |
| sports/control | F1,332 = 5.38 (p = 0.0210) * | 0.016 | 0.638 | ||||||
| CpG sites 22 | F2,332 = 2.78 (p = 0.0962) | 0.008 | 0.383 | ||||||
| addicts/control x CpG sites 22 | F2,332 = 7.55 (p = 0.006) * | 0.022 | 0.782 | ||||||
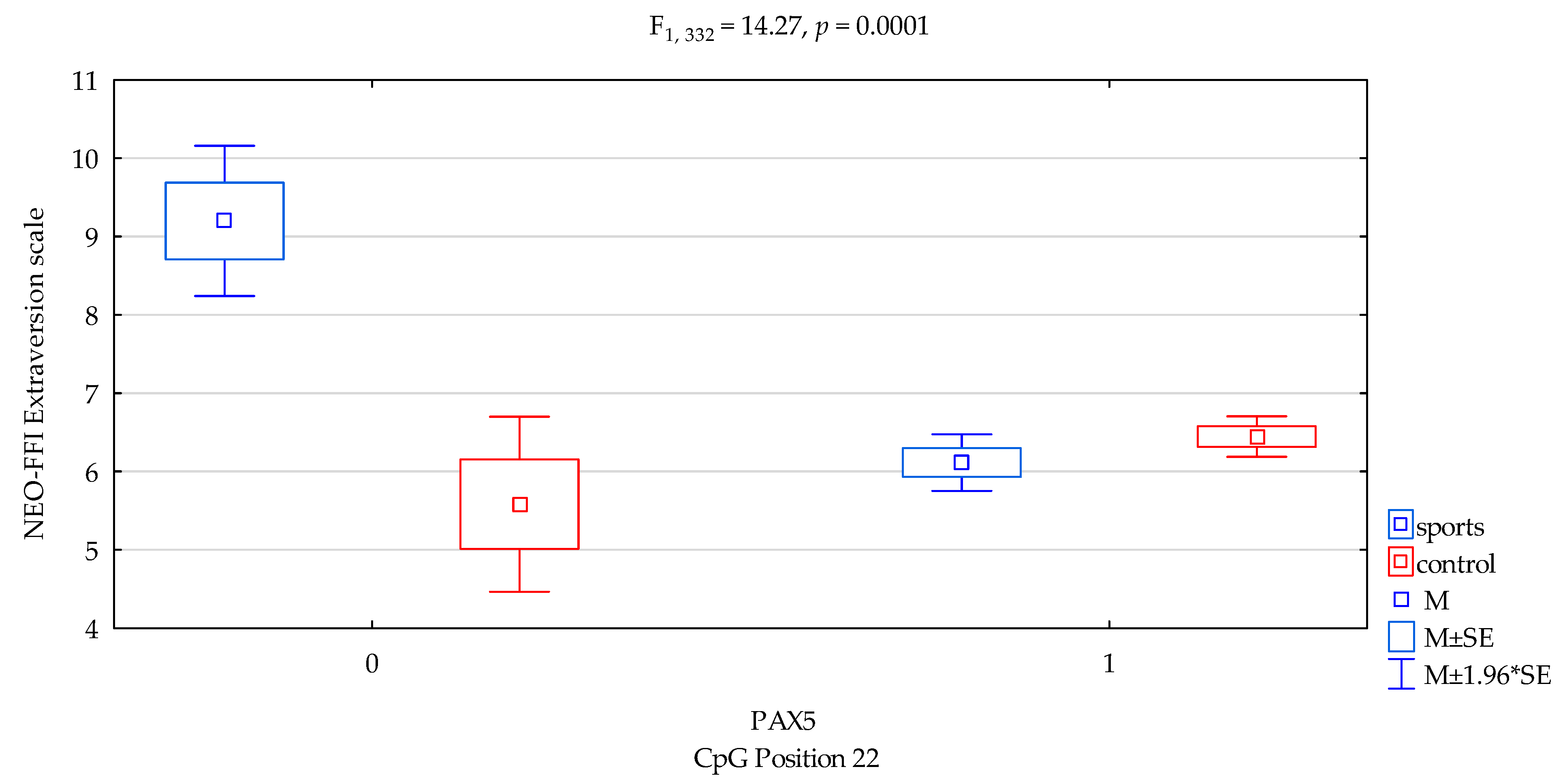
| Extraversion/scale | 6.27 ± 1.89 | 6.41 ± 1.97 | 6.65 ± 2.42 | 6.35 ± 1.92 | F5,332 = 5.001 p = 0.0021 * R2 = 0.0432 | intercept | F1,332 = 685.04 (p < 0.0001) * | 0.673 | 1.000 |
| sports/control | F1,389 = 9.89 (p = 0.0018) * | 0.029 | 0.880 | ||||||
| CpG sites 22 | F2,389 = 4.52 (p = 0.0342) * | 0.013 | 0.563 | ||||||
| addicts/control x CpG sites 22 | F2,389 = 14.27 (p = 0.0001) * | 0.041 | 0.965 | ||||||
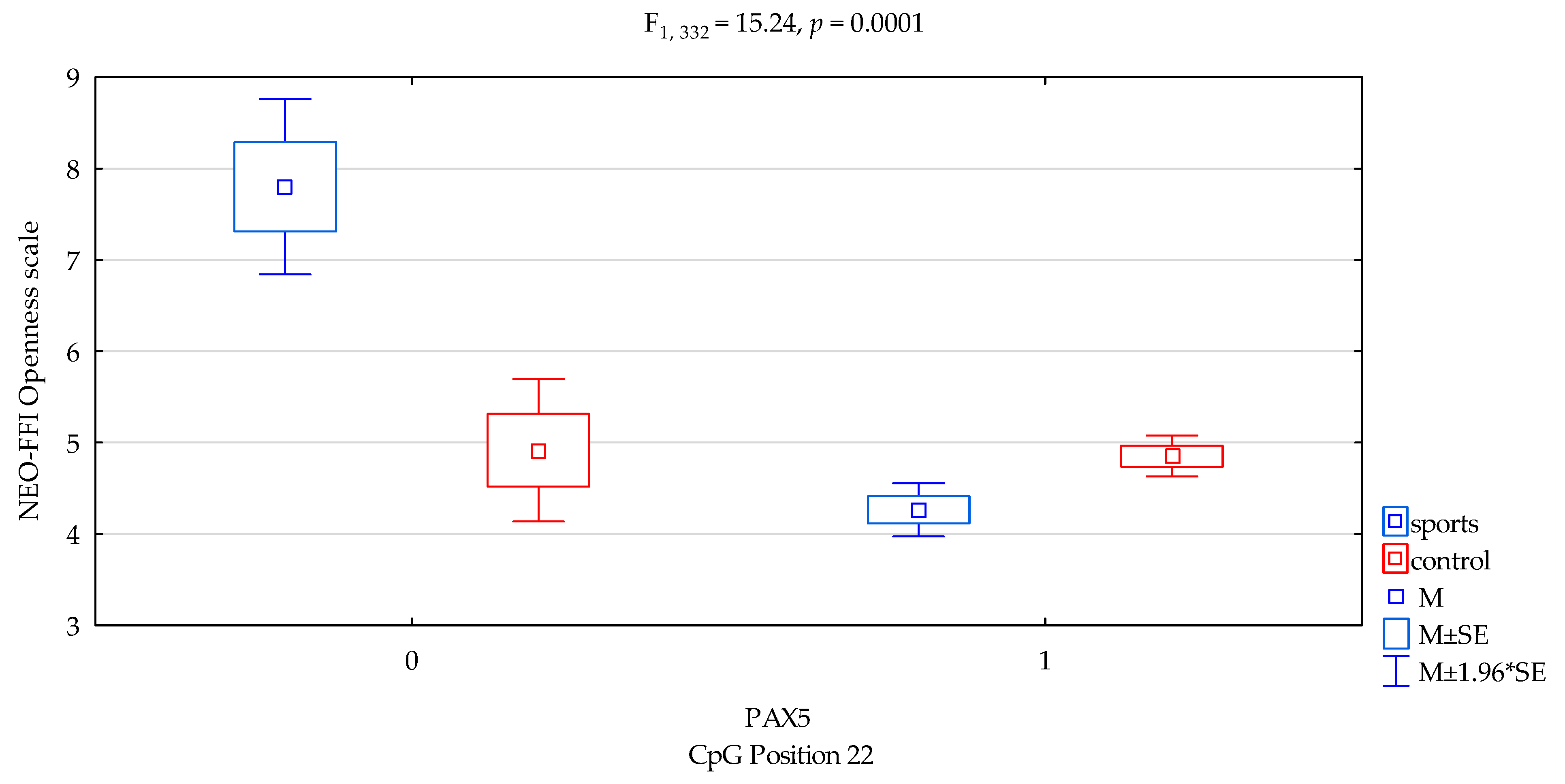
| Openness/scale | 4.44 ± 1.63 | 4.83 ± 1.70 | 5.76 ± 1.86 | 4.68 ± 1.66 | F5,332 = 9.003 p = 0.00001 * R2 = 0.075 | intercept | F1,332 = 602.61 (p < 0.0001) * | 0.644 | 1.000 |
| sports/control | F1,332 = 6.65 (p = 0.0103) * | 0.020 | 0.729 | ||||||
| CpG sites 22 | F2,332 = 16.39 (p = 0.0001) * | 0.047 | 0.981 | ||||||
| addicts/control x CpG sites 22 | F2,332 = 15.24 (p = 0.0001) * | 0.044 | 0.973 | ||||||
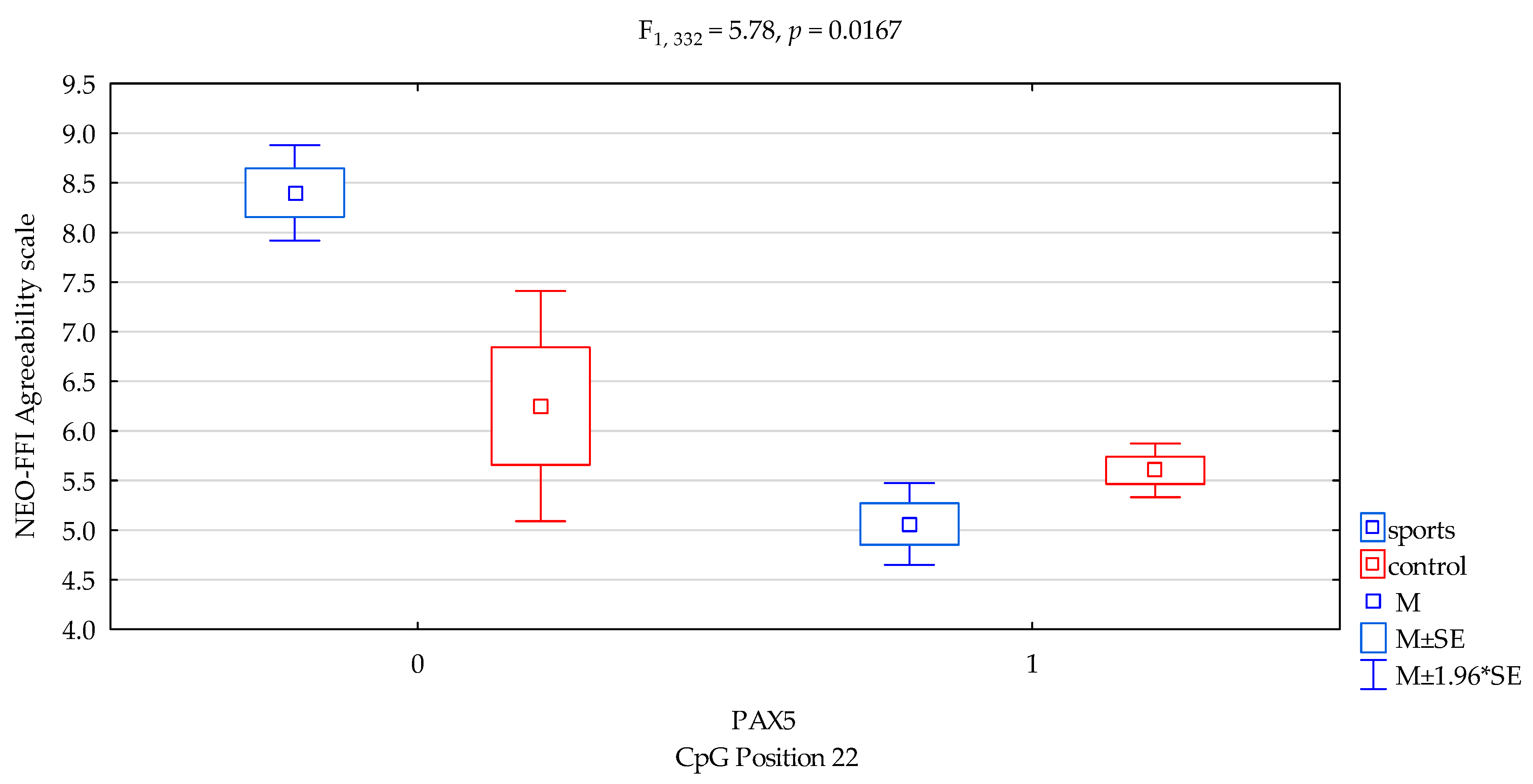
| Agreeability/scale | 5.23 ± 2.13 | 5.65 ± 2.07 | 6.88 ± 1.99 | 5.44 ± 2.07 | F5,332 = 5.500 p = 0.0011 * R2 = 0.047 | intercept | F1,332 = 512.12 (p < 0.0001) * | 0.607 | 1.000 |
| sports/control | F1,332 = 2.07 (p = 0.1509) * | 0.006 | 0.300 | ||||||
| CpG sites 22 | F2,332 = 12.68 (p = 0.0004) * | 0.037 | 0.944 | ||||||
| addicts/control x CpG sites 22 | F2,332 = 5.78 (p = 0.0167) * | 0.017 | 0.669 | ||||||
| DAT1 PAX5 CpG Sites 3 Openness Scale | ||||
|---|---|---|---|---|
| {1} M = 5.27 | {2} M = 4.20 | {3} M = 4.62 | {4} M = 4.87 | |
| Sport; methylation status no {1} | 0.0082 * | 0.2596 | 0.2796 | |
| Sport; methylation status yes {2} | 0.4111 | 0.0026* | ||
| Control; methylation status no {3} | 0.5922 | |||
| Control; methylation status yes {4} | ||||
| DAT1 PAX5 CpG sites 13 Neuroticism scale | ||||
| {1} M = 4.53 | {2} M = 5.36 | {3} M = 4.74 | {4} M = 3.36 | |
| Sport; methylation status no {1} | 0.07151 | 0.4520 | 0.0817 | |
| Sport; methylation status yes {2} | 0.1344 | 0.0069 * | ||
| Control; methylation status no {3} | 0.0314 * | |||
| Control; methylation status yes {4} | ||||
| DAT1 PAX5 CpG sites 22 Neuroticism scale | ||||
| {1} M = 2.40 | {2} M = 4.88 | {3} M = 5.25 | {4} M = 4.64 | |
| Sport; methylation status no {1} | 0.0090 * | 0.0098 * | 0.0166 * | |
| Sport; methylation status yes {2} | 0.5625 | 0.3393 | ||
| Control; methylation status no {3} | 0.3206 | |||
| Control; methylation status yes {4} | ||||
| DAT1 PAX5 CpG sites 22 Extraversion scale | ||||
| {1} M = 9.20 | {2} M = 6.12 | {3} M = 5.58 | {4} M = 6.45 | |
| Sport; methylation status no {1} | 0.0005 * | 0.0004 * | 0.0016 * | |
| Sport; methylation status yes {2} | 0.3642 | 0.1590 | ||
| Control; methylation status no {3} | 0.1288 | |||
| Control; methylation status yes {4} | ||||
| DAT1 PAX5 CpG sites 22 Openness scale | ||||
| {1} M = 7.80 | {2} M = 4.26 | {3} M = 4.92 | {4} M = 4.85 | |
| Sport; methylation status no {1} | 0.0000 * | 0.0010 * | 0.0001 * | |
| Sport; methylation status yes {2} | 0.1911 | 0.0033 * | ||
| Control; methylation status no {3} | 0.8946 | |||
| Control; methylation status yes {4} | ||||
| DAT1 PAX5CpG sites 22 Agreeability scale | ||||
| {1} M = 8.40 | {2} M = 5.06 | {3} M = 6.25 | {4} M = 5.60 | |
| Sport; methylation status no {1} | 0.0004 | 0.0494 * | 0.0027 * | |
| Sport; methylation status yes {2} | 0.0594 | 0.0322 * | ||
| Control; methylation status no {3} | 0.2869 | |||
| Control; methylation status yes {4} | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grzywacz, A.; Chmielowiec, K.; Boroń, A.; Michałowska-Sawczyn, M.; Chmielowiec, J.; Trybek, G.; Mroczek, B.; Leźnicka, K.; Cieszczyk, P.; Masiak, J. Influence of DAT1 Promotor Methylation on Sports Performance. Genes 2021, 12, 1425. https://doi.org/10.3390/genes12091425
Grzywacz A, Chmielowiec K, Boroń A, Michałowska-Sawczyn M, Chmielowiec J, Trybek G, Mroczek B, Leźnicka K, Cieszczyk P, Masiak J. Influence of DAT1 Promotor Methylation on Sports Performance. Genes. 2021; 12(9):1425. https://doi.org/10.3390/genes12091425
Chicago/Turabian StyleGrzywacz, Anna, Krzysztof Chmielowiec, Agnieszka Boroń, Monika Michałowska-Sawczyn, Jolanta Chmielowiec, Grzegorz Trybek, Bożena Mroczek, Katarzyna Leźnicka, Paweł Cieszczyk, and Jolanta Masiak. 2021. "Influence of DAT1 Promotor Methylation on Sports Performance" Genes 12, no. 9: 1425. https://doi.org/10.3390/genes12091425
APA StyleGrzywacz, A., Chmielowiec, K., Boroń, A., Michałowska-Sawczyn, M., Chmielowiec, J., Trybek, G., Mroczek, B., Leźnicka, K., Cieszczyk, P., & Masiak, J. (2021). Influence of DAT1 Promotor Methylation on Sports Performance. Genes, 12(9), 1425. https://doi.org/10.3390/genes12091425









