Descending Dysploidy and Bidirectional Changes in Genome Size Accompanied Crepis (Asteraceae) Evolution
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Amplification and Sequencing
2.3. Sequence Alignment and Phylogenetic Analyses
2.4. Chromosome Preparation and Karyotype Analyses
2.5. Genome Size Measurements
2.6. Ancestral State Reconstructions
3. Results
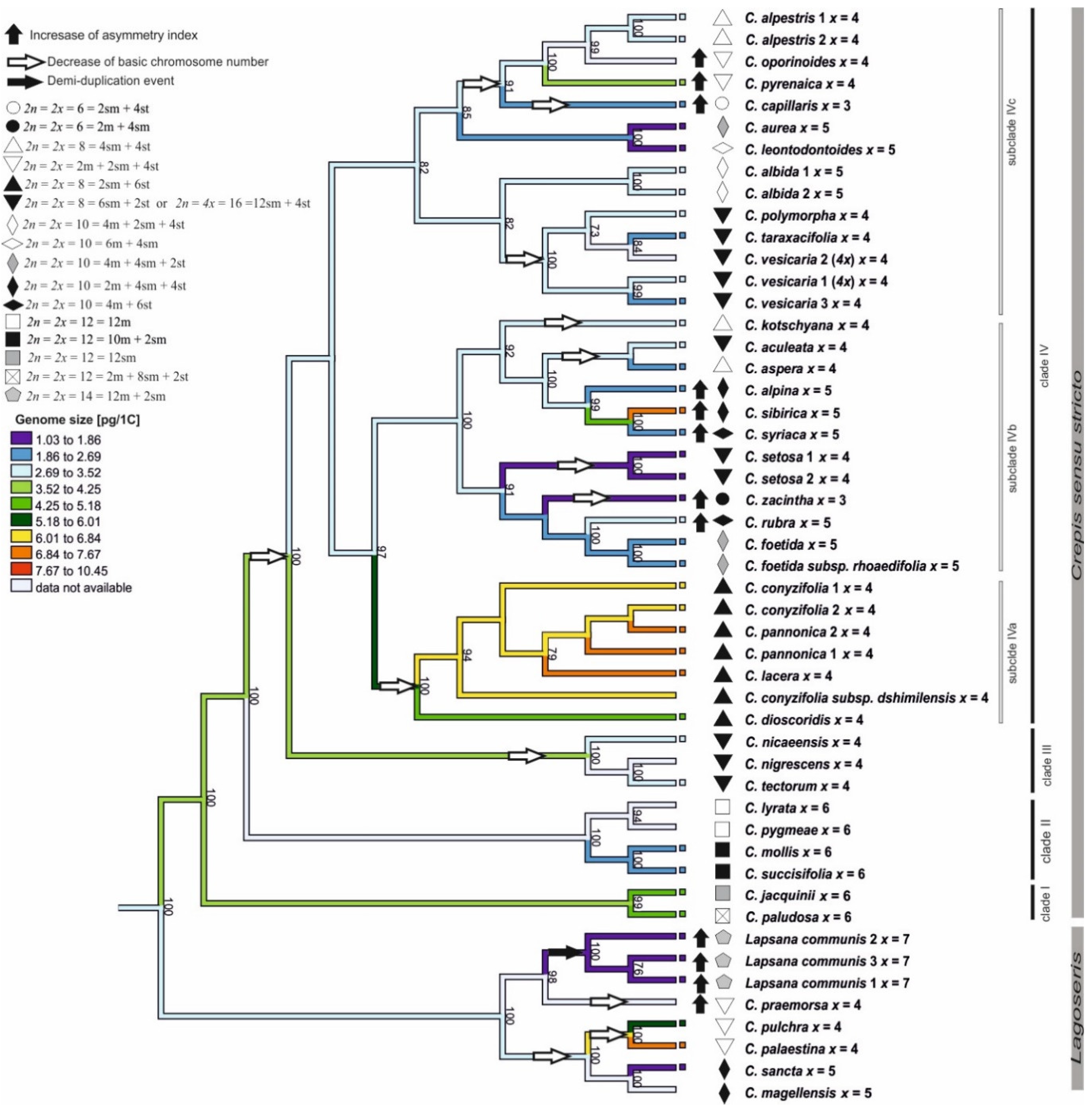
3.1. Phylogenetic Analysis
3.2. Chromosome Number
3.3. Karyotype Structure
3.4. Genome Size
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schubert, I.; Vu, G.T.H. Genome Stability and Evolution: Attempting a Holistic View. Trends Plant Sci. 2016, 21, 749–757. [Google Scholar] [CrossRef]
- Lysák, M.A.; Schubert, I. Mechanisms of Chromosome Rearrangements. In Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer Vienna: Vienna, Austria, 2013; pp. 137–147. [Google Scholar] [CrossRef]
- Coghlan, A.; Eichler, E.E.; Oliver, S.G.; Paterson, A.H.; Stein, L. Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genet. 2005, 21, 673–682. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.K.; Covey, P.A.; Larsen, L.R.; Bedinger, P.; Stack, S.M. Structural differences in chromosomes distinguish species in the tomato clade. Cytogenet. Genome Res. 2010, 129, 24–34. [Google Scholar] [CrossRef]
- Baeza, C.; Finot, V.L.; Ruiz, E. Comparative karyotype analysis of populations in the Alstroemeria presliana Herbert (Alstroemeriaceae) complex in Chile. Genet. Mol. Biol. 2015, 38, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Ambrožová, K.; Mandáková, T.; Bureš, P.; Neumann, P.; Leitch, I.J.; Koblížková, A.; Macas, J.; Lysak, M.A. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Ann. Bot. 2011, 107, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Weiss-Schneeweiss, H.; Schneeweiss, G.M. Karyotype Diversity and Evolutionary Trends in Angiosperms. In Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer Vienna: Vienna, Austria, 2013; pp. 209–230. [Google Scholar] [CrossRef]
- Li, S.F.; Su, T.; Cheng, G.Q.; Wang, B.X.; Li, X.; Deng, C.L.; Gao, W.J. Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants. Genes 2017, 8, 290. [Google Scholar] [CrossRef]
- Funk, V.A.; Susanna, A.; Stuessy, T.F.; Bayer, R.J. Systematics, Evolution, and Biogeography of Compositae; Smithsonian Institution: Washington, DC, USA, 2009. [Google Scholar]
- Babcock, E.B.; Jenkins, J.A. Chromosomes and Phylogeny in Crepis, III: The Relationships of one Hundred and Thirteen Species; University of California Press: Berkeley, CA, USA, 1943. [Google Scholar]
- Enke, N.; Fuchs, J.; Gemeinholzer, B. Shrinking genomes? Evidence from genome size variation in Crepis (Compositae). Plant Biol. 2011, 13, 185–193. [Google Scholar] [CrossRef]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold: London, UK, 1971. [Google Scholar]
- Smocovitis, V.B. The “Plant Drosophila”: E.B. Babcock, the genus “Crepis”, and the evolution of a genetics research program at Berkeley, 1915–1947. Hist. Stud. Nat. Sci. 2009, 39, 300–355. [Google Scholar] [CrossRef]
- Hollingshead, L.; Babcock, E. Chromosomes and Phylogeny in Crepis; University of California Press: Berkeley, CA, USA, 1930; Volume 6, pp. 1–53. [Google Scholar]
- Babcock, E.B.; Cameron, D.R. Chromosomes and Phylogeny in Crepis. II. The Relationships of one Hundred Eight Species; University of California Press: Berkeley, CA, USA, 1934; pp. 287–324. [Google Scholar]
- Da Silva, C.R.M.; Gonzalez-Elizondo, M.S.; Vanzela, A.L.L. Reduction of chromosome number in Eleocharis subarticulata (Cyperaceae) by multiple translocations. Bot. J. Linn. Soc. 2005, 149, 457–464. [Google Scholar] [CrossRef]
- Wan, T.; Zhang, X.; Gregan, J.; Zhang, Y.; Guo, P.; Guo, Y. A dynamic evolution of chromosome in subgenus Potamogeton revealed by physical mapping of rDNA loci detection. Plant Syst. Evol. 2012, 298, 1195–1210. [Google Scholar] [CrossRef]
- Briggs, D.; Walters, S.M. Plant Variation and Evolution, 4th ed.; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar] [CrossRef]
- Babcock, E.B. The Genus Crepis I. The Taxonomy, Phylogeny, Distribution and Evolution of Crepis. The Genus Crepis II. Systematic treatment.; University of California Press: Berkeley, CA, USA, 1947. [Google Scholar]
- Levitsky, G.A. The karyotype in systematics. Bull. Appl. Bot. Genet. Plant Breed. 1931, 27, 220–240. [Google Scholar]
- Enke, N.; Gemeinholzer, B. Babcock revisited: New insights into generic delimitation and character evolution in Crepis L. (Compositae: Cichorieae) from ITS and matK sequence data. Taxon 2008, 57, 756–768. [Google Scholar] [CrossRef]
- Enke, N. Contributions towards a revised infrageneric classification of Crepis (Cichorieae, Compositae). Willdenowia 2010, 39, 229–245. [Google Scholar] [CrossRef]
- Emadzade, K.; Jang, T.-S.; Macas, J.; Kovařík, A.; Novák, P.; Parker, J.; Weiss-Schneeweiss, H. Differential amplification of satellite PaB6 in chromosomally hypervariable Prospero autumnale complex (Hyacinthaceae). Ann. Bot. 2014, 114, 1597–1608. [Google Scholar] [CrossRef]
- Venora, G.; Blangiforti, S.; Frediani, M.; Maggini, F.; Gelati, M.T.; Castiglione, M.R.; Cremonini, R. Nuclear DNA contents, rDNAs, chromatin organization, and karyotype evolution inVicia sect, faba. Protoplasma 2000, 213, 118–125. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Blöch, C.; Weiss-Schneeweiss, H.; Schneeweiss, G.M.; Barfuss, M.H.J.; Rebernig, C.A.; Villaseñor, J.L.; Stuessy, T.F. Molecular phylogenetic analyses of nuclear and plastid DNA sequences support dysploid and polyploid chromosome number changes and reticulate evolution in the diversification of Melampodium (Millerieae, Asteraceae). Mol. Phylogenet. Evol. 2009, 53, 220–233. [Google Scholar] [CrossRef]
- Löytynoja, A.; Goldman, N. webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinform. 2010, 11, 579. [Google Scholar] [CrossRef]
- Collingridge, P.W.; Kelly, S. MergeAlign: Improving multiple sequence alignment performance by dynamic reconstruction of consensus multiple sequence alignments. BMC Bioinform. 2012, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.2, A Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 September 2014).
- Dydak, M.; Kolano, B.; Nowak, T.; Siwinska, D.; Maluszynska, J. Cytogenetic studies of three European species of Centaurea L. (Asteraceae). Hereditas 2009, 146, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Paszko, B. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst. Evol. 2006, 258, 39–48. [Google Scholar] [CrossRef]
- Catalán, P.; Müller, J.; Hasterok, R.; Jenkins, G.; Mur, L.A.J.; Langdon, T.; Betekhtin, A.; Siwinska, D.; Pimentel, M.; López-Alvarez, D. Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 2012, 109, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Sgorbati, S.; Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Temsch, E.M.; Greilhuber, J.; Krisai, R. Genome size in liverworts. Preslia 2010, 82, 63–80. [Google Scholar]
- Lysak, M.A.; Dolezel, J. Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia 1998, 51, 123–132. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Lucretti, S.; Meister, A.; Lysák, M.A.; Nardi, L.; Obermayer, R. Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann. Bot. 1998, 82, 17–26. [Google Scholar] [CrossRef]
- Greilhuber, J.; Dolezel, J.; Lysák, M.A.; Bennett, M.D. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef]
- Glick, L.; Mayrose, I. ChromEvol: Assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Mol. Biol. Evol. 2014, 31, 1914–1922. [Google Scholar] [CrossRef]
- Babcock, E.; Stebbins, G. The American Species of Crepis Their Interrelationships and Distribution as Affected by Polyploidy and Apomixis; Carnegie Institution of Washington: Washington, DC, USA, 1938. [Google Scholar] [CrossRef]
- Babcock, E.B.; Swezy, O. The Chromosomes of Crepis biennis L., and Crepis ciliata C. Koch. Cytologia 1935, 6, 256–265. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Bonnet, T.; Leblois, R.; Rousset, F.; Crochet, P.-A. A reassessment of explanations for discordant introgressions of mitochondrial and nuclear genomes. Evolution 2017, 71, 2140–2158. [Google Scholar] [CrossRef]
- Degnan, J.H.; Rosenberg, N.A. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 2009, 24, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Acosta, C.M.; Premoli, A.C. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Phylogenetics Evol. 2010, 54, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.A.R.; Ree, R.H. Inferring Phylogeny and Introgression using RADseq Data: An Example from Flowering Plants (Pedicularis: Orobanchaceae). Syst. Biol. 2013, 62, 689–706. [Google Scholar] [CrossRef] [PubMed]
- Gaudeul, M.; Gardner, M.F.; Thomas, P.; Ennos, R.A.; Hollingsworth, P.M. Evolutionary dynamics of emblematic Araucaria species (Araucariaceae) in New Caledonia: Nuclear and chloroplast markers suggest recent diversification, introgression, and a tight link between genetics and geography within species. BMC Evol. Biol. 2014, 14, 171. [Google Scholar] [CrossRef]
- Bennett, M.D.; Smith, J.B.; Riley, R. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 274, 227–274. [Google Scholar] [CrossRef] [PubMed]
- Smarda, P.; Knapek, O.; Brezinova, A.; Horova, L.; Grulich, V.; Danihelka, J.; Vesely, P.; Smerda, J.; Rotreklova, O.; Bures, P. Genome sizes and genomic guanine plus cytosine (GC) contents of the Czech vascular flora with new estimates for 1700 species. Preslia 2019, 91, 117–142. [Google Scholar] [CrossRef]
- Bennett, M.D.; Riley, R. Nuclear DNA content and minimum generation time in herbaceous plants. Proc. Biol. Sci. 1972, 181, 109–135. [Google Scholar] [CrossRef]
- Garnatje, T.; Vallès, J.; Garcia, S.; Hidalgo, O.; Sanz, M.; Canela, M.Á.; Siljak-Yakovlev, S. Genome size in Echinops L. and related genera (Asteraceae, Cardueae): Karyological, ecological and phylogenetic implications. Biol. Cell 2004, 96, 117–124. [Google Scholar] [CrossRef]
- Weiss-Schneeweiss, H.; Greilhuber, J.; Schneeweiss, G.M. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am. J. Bot. 2006, 93, 148–156. [Google Scholar] [CrossRef]
- Andrés-Sánchez, S.; Temsch, E.M.; Rico, E.; Montserrat Martínez-Ortega, M. Genome size in Filago L. (Asteraceae, Gnaphalieae) and related genera: Phylogenetic, evolutionary and ecological implications. Plant Syst. Evol. 2013, 299, 331–345. [Google Scholar] [CrossRef]
- Sears, C.J.; Whitton, J. A reexamination of the North American Crepis agamic complex and comparison with the findings of Babcock and Stebbins’ classic biosystematic monograph. Am. J. Bot. 2016, 103, 1289–1299. [Google Scholar] [CrossRef]
- Winterfeld, G.; Paule, J.; Hoffmann, M.H.; Ley, A.; Röser, M. Antagonistic effects of whole-genome duplications and dysploidy on genome sizes in the pantropical monocot family Marantaceae: Consequences in the light of a new molecular phylogeny. Curr. Plant Biol. 2020, 24, 100181. [Google Scholar] [CrossRef]
- Vitales, D.; Álvarez, I.; Garcia, S.; Hidalgo, O.; Nieto Feliner, G.; Pellicer, J.; Vallès, J.; Garnatje, T. Genome size variation at constant chromosome number is not correlated with repetitive DNA dynamism in Anacyclus (Asteraceae). Ann. Bot. 2019, 125, 611–623. [Google Scholar] [CrossRef]
- Pellicer, J.; Kelly, L.J.; Leitch, I.J.; Zomlefer, W.B.; Fay, M.F. A universe of dwarfs and giants: Genome size and chromosome evolution in the monocot family Melanthiaceae. N. Phytol. 2014, 201, 1484–1497. [Google Scholar] [CrossRef]
- Mas de Xaxars, G.; Garnatje, T.; Pellicer, J.; Siljak-Yakovlev, S.; Vallès, J.; Garcia, S. Impact of dysploidy and polyploidy on the diversification of high mountain Artemisia (Asteraceae) and allies. Alp. Bot. 2016, 126, 35–48. [Google Scholar] [CrossRef]
- Fleischmann, A.; Michael, T.P.; Rivadavia, F.; Sousa, A.; Wang, W.; Temsch, E.M.; Greilhuber, J.; Müller, K.F.; Heubl, G. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Ann. Bot. 2014, 114, 1651–1663. [Google Scholar] [CrossRef]
- Winterfeld, G.; Ley, A.; Hoffmann, M.H.; Paule, J.; Röser, M. Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Syst. Evol. 2020, 306, 36. [Google Scholar] [CrossRef]
- Enke, N.; Kunze, R.; Pustahija, F.; Glöckner, G.; Zimmermann, J.; Oberländer, J.; Kamari, G.; Siljak-Yakovlev, S. Genome size shifts: Karyotype evolution in Crepis section Neglectoides (Asteraceae). Plant Biol. 2015, 17, 775–786. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet. Genome Res. 2005, 110, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L. Modes, rates and mechanisms of local genomic change in flowering plants: Tempest in a pea plot. In Plant Species-Level Systematics: New Perspectives on Pattern & Process; Bakker, F.T., Chatrou, L.W., Gravendeel, B., Pelser, P.B., Eds.; A.R.G. Gantner Verlag: Ruggell, Liechtenstein, 2005; p. 348. [Google Scholar]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Ellison, N.W.; Liston, A.; Steiner, J.J.; Williams, W.M.; Taylor, N.L. Molecular phylogenetics of the clover genus (Trifolium—Leguminosae). Mol. Phylogenetics Evol. 2006, 39, 688–705. [Google Scholar] [CrossRef] [PubMed]
- Cerbah, M.; Coulaud, J.; Brown, S.; Siljak-yakovlev, S. Evolutionary DNA variation in the genus Hypochaeris. Heredity 1999, 82, 261–266. [Google Scholar] [CrossRef]
- Watanabe, K.; Yahara, T.; Denda, T.; Kosuge, K. Chromosomal Evolution in the Genus Brachyscome (Asteraceae, Astereae): Statistical Tests Regarding Correlation Between Changes in Karyotype and Habit Using Phylogenetic Information. J. Plant. Res. 1999, 112, 145–161. [Google Scholar] [CrossRef]
- Shan, F.; Yan, G.; Plummer, J.A. Karyotype evolution in the genus Boronia (Rutaceae). Bot. J. Linn. Soc. 2003, 142, 309–320. [Google Scholar] [CrossRef]
- Mandáková, T.; Lysak, M.A. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 2008, 20, 2559–2570. [Google Scholar] [CrossRef] [PubMed]
- Baltisberger, M.; Hörandl, E. Karyotype evolution supports the molecular phylogeny in the genus Ranunculus (Ranunculaceae). Perspect. Plant Ecol. Evol. Syst. 2016, 18, 1–14. [Google Scholar] [CrossRef][Green Version]
- Lysak, M.A.; Koch, M.A. Phylogeny, Genome, and Karyotype Evolution of Crucifers (Brassicaceae). In Genetics and Genomics of the Brassicaceae. Plant Genetics and Genomics: Crops and Models; Schmidt, R., Bancroft, I., Eds.; Springer: New York, NY, USA, 2011. [Google Scholar]
- Lysak, M.A.; Berr, A.; Pecinka, A.; Schmidt, R.; McBreen, K.; Schubert, I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 2006, 103, 5224–5229. [Google Scholar] [CrossRef]
- Lusinska, J.; Betekhtin, A.; Lopez-Alvarez, D.; Catalan, P.; Jenkins, G.; Wolny, E.; Hasterok, R. Comparatively Barcoded Chromosomes of Brachypodium Perennials Tell the Story of Their Karyotype Structure and Evolution. Int. J. Mol. Sci. 2019, 20, 5557. [Google Scholar] [CrossRef]
- Stebbins, G.L. Chromosomal Variation and Evolution. Science 1966, 152, 1463–1469. [Google Scholar] [CrossRef]
- Wallace, H.; Sparkes, C.A.; Maden, M. Nuclear DNA content of three Crepis species. Heredity 1972, 29, 367–373. [Google Scholar] [CrossRef][Green Version]
- Dimitrova, D.; Greilhuber, J. Karyotype and DNA-content evolution in ten species of Crepis (Asteraceae) distributed in Bulgaria. Bot. J. Linn. Soc. 2000, 132, 281–297. [Google Scholar] [CrossRef][Green Version]




| Taxon | Collection Details | Voucher | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|
| trnK-rps16 | rpl32-trnL | trnT-psbD | ndhC-trnV | nrITS | |||
| Crepis s.s. | |||||||
| Crepis aculeata Boiss. | BGT 38 | KTU154623 | MT234677 | MT234860 | MT234738 | MT234799 | MN549102 |
| C. acuminata Nutt. | USDA W6 40086 N 41°20′31.217″ W 119°50′57.293″ | - | MT234678 | MT234861 | MT234739 | MT234800 | MN549103 |
| C. albida (1) Vill. | UGA 233 | - | MT234679 | MT234862 | MT234740 | MT234801 | MN549111 |
| C. albida (2) Vill. | JBI 2 | KTU157715 | MT234680 | MT234863 | MT234741 | MT234802 | MN549112 |
| C. alpestris (1) (Jacq.) Tausch | BGUG N49 | KTU157712 | MT234681 | MT234864 | MT234742 | MT234803 | MN549104 |
| C. alpestris (2) (Jacq.) Tausch | BGBD 708 N 47°46′43.18″ E 13°26′01.16″ | KTU157714 | MT234682 | MT234865 | MT234743 | MT234804 | MN549105 |
| C. alpina L. | USDA PI 274367 | KTU154609 | MT234683 | MT234866 | MT234744 | MT234805 | MN549106 |
| C. aspera L. | LBG 006722 | KTU157716 | MT234685 | MT234868 | MT234746 | MT234807 | MN549108 |
| C. atribarba A.Heller | USDA W6 36843 | - | MT234686 | MT234869 | MT234747 | MT234808 | MN549109 |
| C. aurea (L.) Cass. | USDA PI 312843 | KTU157719 | MT234687 | MT234870 | MT234748 | MT234809 | MN549110 |
| C. biennis L. 1 | BGBD 656 N 51°11′16.44″ E 10°3′4572″ | KTU154629 | MT234688 | MT234871 | MT234749 | MT234810 | MN549113 |
| C. biennis L. 2 | Wołosate, Poland N 49°03′29.60″ E 22°42′08.92″ | KTU157728 | MT234689 | MT234872 | MT234750 | MT234811 | MN549114 |
| C. capillaris Wallr. | BGGU 335 | KTU154610 | MT234691 | MT234874 | MT234752 | MT234813 | MN549116 |
| C. conyzifolia (Gouan) A.Kern. | GBA 462 | KTU157720 | MT234692 | MT234875 | MT234753 | MT234814 | MN549117 |
| C. conyzifolia (Gouan) A.Kern. | UGA 236 | - | MT234693 | MT234876 | MT234754 | MT234815 | MN549118 |
| C. conyzifolia subsp. dshimilensis (K.Koch) Lamond | GBBG | - | MT234714 | MT234897 | MT234775 | MT234836 | MN549132 |
| C. dioscoridis L. | IPK CRE2 | KTU154619 | MT234694 | MT234877 | MT234755 | MT234816 | MN549119 |
| C. foetida L. | USDA PI 296071 | KTU154612 | MT234695 | MT234878 | MT234756 | MT234817 | MN549120 |
| C. foetida subsp. rhoaedifolia (M.Bieb.) Celak. | HBBH 1734 | KTU154614 | MT234718 | MT234901 | MT234779 | MT234840 | MN549137 |
| C. intermedia A.Gray | USDA W6 52751 N 39°48′15.984″ W 119°56′2.867″ | - | MT234696 | MT234879 | MT234757 | MT234818 | MN549121 |
| C. jacquinii Tausch | Sarnia Skała Tatra Mts, Poland N 49°15′52.77″ E 19°56′30.36″ | KTU159736 | MT234697 | MT234880 | MT234758 | MT234819 | MT234671 |
| C. kotschyana Boiss. | USDA PI 310392 | KTU164608 | MT234698 | MT234881 | MT234759 | MT234820 | MN549122 |
| C. lacera Ten. | BGMN | KTU159735 | MT234699 | MT234882 | MT234760 | MT234821 | MT234672 |
| C. leontodontoides All. | BGDG 658 N 42°8′43.044″ E 8°59′32.964″ | KTU154631 | MT234700 | MT234883 | MT234761 | MT234822 | MN549123 |
| C. lyrata (L.) Froel. | SSBG | - | MT234701 | MT234884 | MT234762 | MT234823 | MT234673 |
| C. modocensis Greene | USDA W6 49189 N 45°3′19.368″ W 109°10′4.079″ | - | MT234703 | MT234886 | MT234764 | MT234825 | MN549124 |
| C. mollis Asch. | Sławków, Poland N 50°17′45.90″ E 19°16′59.06″ | KTU154630 | MT234704 | MT234887 | MT234765 | MT234826 | MN549125 |
| C. nicaeensis Balb. | BGEU | KTU157730 | MT234705 | MT234888 | MT234766 | MT234827 | MT234675 |
| C. nigrescens Pohle | HUM | MW0553775 | MT234706 | MT234889 | MT234767 | MT234828 | MN549126 |
| C. occidentalis Nutt. | USDA W6 45275 N 43°19′50.7″ W 117°11′0.708″ | - | MT234707 | MT234890 | MT234768 | MT234829 | MN549127 |
| C. oporinoides Boiss. ex Froel. | ABGL 1516 | KTU154622 | MT234684 | MT234867 | MT234745 | MT234806 | MN549107 |
| C. paludosa Moench | Sławków, Poland N 50°18′07.51″ E 19°21′19.10″ | KTU154625 | MT234709 | MT234892 | MT234770 | MT234831 | MN549128 |
| C. pannonica (Jacq.) K.Koch 1 | BGBD 256-01-00-14 N 48°21′58.54″ E 16°25′06.13″ | KTU154627 | MT234710 | MT234893 | MT234771 | MT234832 | MN549130 |
| C. pannonica (Jacq.) K.Koch 2 | BGEU | KTU157729 | MT234711 | MT234894 | MT234772 | MT234833 | MT234676 |
| C. polymorpha Pourr | JBN 149 | KTU157725 | MT234713 | MT234896 | MT234774 | MT234835 | MN549131 |
| C. pontana Dalla Torre | ABGL 235 | KTU154624 | MT234690 | MT234873 | MT234751 | MT234812 | MN549115 |
| C. pygmaea L. | UGA 239 | KTU157722 | MT234716 | MT234899 | MT234777 | MT234838 | MN549135 |
| C. pyrenaica (L.) Greuter | BGBD 1010 N 42°52′22.44″ W 0°26′56.616″ | KTU154621 | MT234717 | MT234900 | MT234778 | MT234839 | MN549136 |
| C. rubra L. | BGK 364 | KTU154607 | MT234719 | MT234902 | MT234780 | MT234841 | MN549138 |
| C. setosa Haller f. 1 | HBUR 1275 | KTU154620 | MT234722 | MT234905 | MT234783 | MT234844 | MN549140 |
| C. setosa Haller f. 2 | IPK CRE20 | KTU157713 | MT234721 | MT234904 | MT234782 | MT234843 | MN549141 |
| C. sibirica L. | BGBD 738 N 60°21′34.37″ E 59°11′22.58″ | KTU157721 | MT234723 | MT234906 | MT234784 | MT234845 | MN549142 |
| C. succisifolia Tausch | Rędziny, Poland N 50°49′08.66″ E 15°55′55.27″ | KTU154656 | MT234724 | MT234907 | MT234785 | MT234846 | MN549143 |
| C. syriaca (Bornm.) Babc. & Navashin | KEW 0129064 | KTU154615 | MT234725 | MT234908 | MT234786 | MT234847 | MN549144 |
| C. taraxacifolia Thuill. | BGGU 347 | KTU157723 | MT234726 | MT234909 | MT234787 | MT234848 | MN549145 |
| C. tectorum L. | Ustroń, Poland N 49°43′14.68″ E 18°49′29.11″ | KTU157717 | MT234727 | MT234910 | MT234788 | MT234849 | MN549146 |
| C. vesicaria L. 3 | OBUP | KTU157724 | MT234730 | MT234913 | MT234791 | MT234852 | MN549149 |
| C. vesicaria L. 2 | BGBD 918 N 39°34′24.816″ E 2°38′42.648″ | KTU157726 | MT234729 | MT234912 | MT234790 | MT234851 | MN549148 |
| C. veiscaria L. 1 | BGBD 1014 | KTU154616 | MT234728 | MT234911 | MT234789 | MT234850 | MN549147 |
| C. zacintha (L.) Loisel. | BGT 92 | KTU154606 | MT234731 | MT234914 | MT234792 | MT234853 | MN549150 |
| Lagoseris | |||||||
| C. magellensis F.Conti & Uzunov | BGMN | KTU157727 | MT234702 | MT234885 | MT234763 | MT234824 | MT234674 |
| C. palaestina Bornm. | BGGU 335 | KTU154611 | MT234708 | MT234891 | MT234769 | MT234830 | MN549129 |
| C. pulchra L. | BGGU 341 | KTU154648 | MT234712 | MT234895 | MT234773 | MT234834 | MN549134 |
| C. preamorsa (L.) Tausch | BGBD 662 N 60°18′21.29″ E 10°35′20.76″ | KTU154628 | MT234715 | MT234898 | MT234776 | MT234837 | MN549133 |
| C. sancta (L.) Bornm. | BGUK 104 | KTU154613 | MT234720 | MT234903 | MT234781 | MT234842 | MN549139 |
| Lapsana communis L. 1 | KEW 0018568 | KTU154617 | MT234733 | MT234916 | MT234794 | MT234855 | MN549151 |
| L. communis L. 2 | Rogoźnik Poland N 50°24′03.43″; E 19°01′59.96″ | KTU157708 | MT234734 | MT234917 | MT234795 | MT234856 | MN549152 |
| L. communis L. 3 | Rogoźnik Poland N 50°23′50.22″; E 19°01′48.50″ | KTU157709 | MT234735 | MT234918 | MT234796 | MT234857 | MN549153 |
| Outgroup | |||||||
| Lactuca serriola L. | Strzyżowice Poland N 50°23′33.47″ E 19°04′03.14″ | KTU157718 | MT234732 | MT234915 | MT234793 | MT234854 | MN549156 |
| Picris hieracioides L. | Jaworzno Poland N 50°13′31.43″ E 19°16′28.63″ | KTU157710 | MT234736 | MT234919 | MT234797 | MT234858 | MN549154 |
| Sonchus oleraceus L. | Ustroń, Poland N 49°42′58.97″ E 18°47′53.44″ | KTU157711 | MT234737 | MT234920 | MT234798 | MT234859 | MN549157 |
| Species | Karyotype Formula * | Asymmetry Index (AI) | Genome Size pg/1C ± SD | Internal Standard |
|---|---|---|---|---|
| Crepis s.s. | ||||
| Crepis aculeata Boiss. | 2n = 2x = 8 = 6sm + 2st | 1.38 | 2.89 ± 0.04 | Pisum sativum |
| C. albida Vill. (1) | 2n = 2x = 10 = 4m + 2sm + 4st | 7.07 | 3.08 ± 0.03 | P. sativum |
| C. albida Vill. (2) | 2n = 2x = 10 = 4m + 2sm + 4st | 7.39 | - | - |
| C. alpestris (Jacq.) Tausch (1) | 2n = 2x = 8 = 4sm + 4st | 3.96 | 2.99 ± 0.03 | P. sativum |
| C. alpestris (Jacq.) Tausch (2) | 2n = 2x = 8 = 4sm + 4st | 2.52 | - | - |
| C. alpina L. | 2n = 2x = 10 = 2m + 4sm + 4st | 13.47 | 2.20 ± 0.05 | Zea mays |
| C. aspera L. | 2n = 2x = 8 = 4sm + 4st | 4.24 | 2.15 ± 0.02 | Lycopersicon pseudocapsicum |
| C. aurea (L.) Cass. | 2n = 2x = 10 = 4m + 4sm + 2st | 5.93 | 1.63 ± 0.10 | Lycopersicon esculentum |
| C. biennis L. (2) | 2n = 8x = 40 | - | 10.45 ± 0.33 | P. sativum |
| C. capillaris Wallr. | 2n = 2x = 6 = 2sm + 4st | 20.15 | 2.07 ± 0.08 | Z. mays |
| C. conyzifolia (Gouan) A.Kern. (1) | 2n = 2x = 8 = 2sm + 6st | 3.74 | 6.08 ± 0.16 | P. sativum |
| C. conyzifolia (Gouan) A.Kern. (2) | 2n = 2x = 8 = 2sm + 6st | 2.06 | - | - |
| C. conyzifolia subsp. dshimilensis (K.Koch) Lamond | 2n = 2x = 8 = 2sm + 6st | 3.67 | - | - |
| C. dioscoridis L. | 2n = 2x = 8 = 2sm + 6st | 4.74 | 4.58 ± 0.13 | Secale cereale |
| C. foetida L. | 2n = 2x = 10 = 4m + 4sm + 2st | 11.17 | 2.03 ± 0.08 | L. pseudocapsicum |
| C. foetida subsp. rhoaedifolia (M.Bieb.) Celak. | 2n = 2x = 10 = 4m + 4sm + 2st | 10.92 | 2.17 ± 0.02 | L. pseudocapsicum |
| C. jacquinii Tausch | 2n = 2x = 12 = 12sm | 5.93 | 5.12 ± 0.05 | S. cereale |
| C. kotschyana Boiss. | 2n = 2x = 8 = 4sm + 4st | 5.54 | 2.92 ± 0.08 | P. sativum |
| C. lacera Ten. | 2n = 2x = 8 = 2sm + 6st | 5.54 | 7.46 ± 0.10 | P. sativum |
| C. leontodontoides All. | 2n = 2x = 10 = 6m + 4sm | 5.37 | 1.06 ± 0.03 | Brachypodium hybridum |
| C. lyrata (L.) Froel. | 2n = 2x = 12 = 12m | 5.72 | - | - |
| C. mollis Asch. | 2n = 2x = 12 = 10m + 2sm | 9.57 | 2.53 ± 0.04 | L. pseudocapsicum |
| C. nicaeensis Balb. | 2n = 2x = 8 = 6sm + 2st | 3.74 | 3.17 ± 0.02 | P. sativum |
| C. nigrescens Pohle | 2n = 2x = 8 = 6sm + 2st | 3.45 | - | - |
| C. oporinoides Boiss. ex Froel. | 2n = 2x = 8 = 2m + 2sm + 4st | 16.56 | - | - |
| C. paludosa Moench | 2n = 2x = 12 = 2m + 8sm +2st | 10.92 | 4.53 ± 0.19 | S. cereale |
| C. pannonica (Jacq.) K.Koch (1) | 2n = 2x = 8 = 2sm + 6st | 6.52 | 7.27 ± 0.06 | P. sativum |
| C. pannonica (Jacq.) K.Koch (2) | 2n = 2x = 8 = 2sm + 6st | 6.41 | - | - |
| C. polymorpha Pourr | 2n = 2x = 8 = 6sm +2st | 1.61 | 3.13 ± 0.02 | P. sativum |
| C. pygmeae L. | 2n = 2x = 12 = 12m | 7.53 | - | - |
| C. pyrenaica (L.) Greuter | 2n = 2x = 8 = 2m + 2sm + 4st | 11.66 | 3.54 ± 0.05 | S. cereale |
| C. rubra L. | 2n = 2x = 10 = 4m + 6st | 12.85 | 2.86 ± 0.07 | P. sativum |
| C. setosa Haller f. (1) | 2n = 2x = 8 = 6sm + 2st | 10.66 | 1.67 ± 0.02 | L. esculentum |
| C. setosa Haller f. (2) | 2n = 2x = 8 = 6sm + 2st | 7.30 | - | - |
| C. sibirica L. | 2n = 2x = 10 = 2m + 4sm + 4st | 14.44 | 6.98 ± 0.04 | P. sativum |
| C. succisifolia Tausch | 2n = 2x = 12 = 10m + 2sm | 10.77 | 2.34 ± 0.05 | L. pseudocapsicum |
| C. syriaca (Bornm.) Babc. & Navashin | 2n = 2x = 10 = 4m + 6st | 75.39 | 2.39 ± 0.28 | L. pseudocapsicum |
| C. taraxacifolia Thuill. | 2n = 2x = 8 = 6sm + 2st | 1.01 | 2.47 ± 0.58 | P. sativum |
| C. tectorum L. | 2n = 2x = 8 = 6sm + 2st | 2.308 | 3.06 ± 0.05 | P. sativum |
| C. vesicaria L. (3) | 2n = 2x = 8 = 6sm + 2st | 2.38 | 2.43 ± 0.03 | L. pseudocapsicum |
| C. vesicaria L. (1) | 2n = 4x = 16 = 12sm + 4st | 2.68 | 2.78 ± 0.09 | P. sativum |
| C. vesicaria L. (2) | 2n = 4x = 16 = 12sm + 4st | 3.05 | - | - |
| C. zacintha (L.) Loisel. | 2n = 2x = 6 = 2m+ 4sm | 18.61 | 1.03 ± 0.02 | Brachypodium hybridum |
| Lagoseris | ||||
| C. magellensis F.Conti & Uzunov | 2n = 2x = 10 = 2m + 4sm + 4st | 5.26 | - | - |
| C. palaestina Bornm. | 2n = 2x = 8 = 2m + 2sm + 4st | 10.45 | 7.05 ± 0.19 | P. sativum |
| C. praemorsa (L.) Tausch | 2n = 2x = 8 = 2m + 2sm + 4st | 27.15 | - | - |
| C. pulchra L. | 2n = 2x = 8 = 2m + 2sm + 4st | 10.2 | 5.59 ± 0.12 | P. sativum |
| C. sancta (L.) Bornm. | 2n = 2x = 10 = 2m + 4sm + 4st | 5.37 | 1.60 ± 0.02 | Z. mays |
| Lapsana communis L. (1) | 2n = 2x = 14 = 12 m + 2sm | 12.06 | 1.22 ± 0.04 | Z. mays |
| L. communis L. (2) | 2n = 2x = 14 = 12 m + 2sm | 12.70 | - | - |
| L. communis L. (3) | 2n = 2x = 14 = 12 m + 2sm | 13.67 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senderowicz, M.; Nowak, T.; Rojek-Jelonek, M.; Bisaga, M.; Papp, L.; Weiss-Schneeweiss, H.; Kolano, B. Descending Dysploidy and Bidirectional Changes in Genome Size Accompanied Crepis (Asteraceae) Evolution. Genes 2021, 12, 1436. https://doi.org/10.3390/genes12091436
Senderowicz M, Nowak T, Rojek-Jelonek M, Bisaga M, Papp L, Weiss-Schneeweiss H, Kolano B. Descending Dysploidy and Bidirectional Changes in Genome Size Accompanied Crepis (Asteraceae) Evolution. Genes. 2021; 12(9):1436. https://doi.org/10.3390/genes12091436
Chicago/Turabian StyleSenderowicz, Magdalena, Teresa Nowak, Magdalena Rojek-Jelonek, Maciej Bisaga, Laszlo Papp, Hanna Weiss-Schneeweiss, and Bozena Kolano. 2021. "Descending Dysploidy and Bidirectional Changes in Genome Size Accompanied Crepis (Asteraceae) Evolution" Genes 12, no. 9: 1436. https://doi.org/10.3390/genes12091436
APA StyleSenderowicz, M., Nowak, T., Rojek-Jelonek, M., Bisaga, M., Papp, L., Weiss-Schneeweiss, H., & Kolano, B. (2021). Descending Dysploidy and Bidirectional Changes in Genome Size Accompanied Crepis (Asteraceae) Evolution. Genes, 12(9), 1436. https://doi.org/10.3390/genes12091436






