HAUSP Is a Key Epigenetic Regulator of the Chromatin Effector Proteins
Abstract
:1. Introduction
2. Role of HAUSP in the Maintenance of DNA Methylation
2.1. HAUSP-Dependent Regulation of DNMT1 by Ubiquitination
2.2. HAUSP-Dependent Regulation of DNMT1 by Acetylation
3. Role of HAUSP in the Regulation of Histone Post-Translational Modifications
3.1. Role of HAUSP in the Regulation of Histone Monoubiquitination
3.2. Role of HAUSP in the Regulation of Histone Acetylation
3.3. Role of HAUSP in the Regulation of Histone Methylation
3.4. Role of HAUSP in the UHRF1-Mediated Readout of Histone H3 Lysine 9 Methylation
4. HAUSP Inhibitors as Promising Anticancer Agents
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rawat, R.; Starczynowski, D.T.; Ntziachristos, P. Nuclear deubiquitination in the spotlight: The multifaceted nature of USP7 biology in disease. Curr. Opin. Cell Biol. 2019, 58, 85–94. [Google Scholar] [CrossRef]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair 2017, 56, 92–101. [Google Scholar] [CrossRef]
- Liu, J.; Shaik, S.; Dai, X.; Wu, Q.; Zhou, X.; Wang, Z.; Wei, W. Targeting the ubiquitin pathway for cancer treatment. Biochim. Biophys. Acta 2015, 1855, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Yerlikaya, A.; Kanbur, E.; Stanley, B.A.; Tümer, E. The Ubiquitin-Proteasome Pathway and Epigenetic Modifications in Cancer. Anti-Cancer Agents Med. Chem. 2021, 21, 20–32. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [Green Version]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 310–319. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Mennerich, D.; Kubaichuk, K.; Kietzmann, T. DUBs, Hypoxia, and Cancer. Trends Cancer 2019, 5, 632–653. [Google Scholar] [CrossRef] [Green Version]
- Poondla, N.; Chandrasekaran, A.P.; Kim, K.S.; Ramakrishna, S. Deubiquitinating enzymes as cancer biomarkers: New therapeutic opportunities? BMB Rep. 2019, 52, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Farshi, P.; Deshmukh, R.R.; Nwankwo, J.O.; Arkwright, R.T.; Cvek, B.; Liu, J.; Dou, Q.P. Deubiquitinases (DUBs) and DUB inhibitors: A patent review. Expert Opin. Ther. Pat. 2015, 25, 1191–1208. [Google Scholar] [CrossRef] [Green Version]
- McClurg, U.L.; Robson, C.N. Deubiquitinating enzymes as oncotargets. Oncotarget 2015, 6, 9657–9668. [Google Scholar] [CrossRef] [Green Version]
- Song, M.S.; Salmena, L.; Carracedo, A.; Egia, A.; Lo-Coco, F.; Teruya-Feldstein, J.; Pandolfi, P.P. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 2008, 455, 813–817. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, J.; Chen, C.; Yuan, H.; Wen, X.; Sun, H. USP7, Target Validation and Drug Discovery for Cancer Therapy. Med. Chem. 2018, 14, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Chakraborty, D.; Basu, M.; Ghosh, M.K. Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct. Target. Ther. 2018, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.D.; Dong, Y.F.; Niu, W.X.; Niu, C.S. HAUSP promoted the growth of glioma cells in vitro and in vivo via stabilizing NANOG. Pathol. Res. Pract. 2020, 216, 152883. [Google Scholar] [CrossRef]
- Tavana, O.; Li, D.; Dai, C.; Lopez, G.; Banerjee, D.; Kon, N.; Chen, C.; Califano, A.; Yamashiro, D.J.; Sun, H.; et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 2016, 22, 1180–1186. [Google Scholar] [CrossRef] [Green Version]
- Pozhidaeva, A.; Bezsonova, I. USP7, Structure, substrate specificity, and inhibition. DNA Repair 2019, 76, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Holowaty, M.N.; Sheng, Y.; Nguyen, T.; Arrowsmith, C.; Frappier, L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 2003, 278, 47753–47761. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Martinez, C.A.; Arcipowski, K.M.; Zhu, Y.; Gutierrez-Diaz, B.T.; Wang, K.K.; Johnson, M.R.; Volk, A.G.; Wang, F.; Wu, J.; et al. USP7 Cooperates with NOTCH1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 222–239. [Google Scholar] [CrossRef] [Green Version]
- Shan, H.; Li, X.; Xiao, X.; Dai, Y.; Huang, J.; Song, J.; Liu, M.; Yang, L.; Lei, H.; Tong, Y.; et al. USP7 deubiquitinates and stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal Transduct. Target. Ther. 2018, 3, 29. [Google Scholar] [CrossRef] [Green Version]
- Novellasdemunt, L.; Foglizzo, V.; Cuadrado, L.; Antas, P.; Kucharska, A.; Encheva, V.; Snijders, A.P.; Li, V.S.W. USP7 Is a Tumor-Specific WNT Activator for APC-Mutated Colorectal Cancer by Mediating β-Catenin Deubiquitination. Cell Rep. 2017, 21, 612–627. [Google Scholar] [CrossRef] [Green Version]
- Franqui-Machin, R.; Hao, M.; Bai, H.; Gu, Z.; Zhan, X.; Habelhah, H.; Jethava, Y.; Qiu, L.; Frech, I.; Tricot, G.; et al. Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J. Clin. Investig. 2018, 128, 2877–2893. [Google Scholar] [CrossRef] [PubMed]
- Tavana, O.; Sun, H.; Gu, W. Targeting HAUSP in both p53 wildtype and p53-mutant tumors. Cell Cycle 2018, 17, 823–828. [Google Scholar] [CrossRef] [Green Version]
- Brooks, C.L.; Li, M.; Hu, M.; Shi, Y.; Gu, W. The p53–Mdm2–HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 2007, 26, 7262–7266. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Ghosh, M.K. HAUSP, a novel deubiquitinase for Rb-MDM2 the critical regulator. FEBS J. 2014, 281, 3061–3078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meulmeester, E.; Pereg, Y.; Shiloh, Y.; Jochemsen, A.G. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle 2005, 4, 1166–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. 2016, 11, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.C.; Jochemsen, A.G. Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Commun. 2005, 331, 750–760. [Google Scholar] [CrossRef]
- Melo, A.N.; Eischen, C.M. Protecting the genome from mdm2 and mdmx. Genes Cancer 2012, 3, 283–290. [Google Scholar] [CrossRef]
- Li, Y.; Reverter, D. Molecular Mechanisms of DUBs Regulation in Signaling and Disease. Int. J. Mol. Sci. 2021, 22, 986. [Google Scholar] [CrossRef] [PubMed]
- Alhosin, M.; Sharif, T.; Mousli, M.; Etienne-Selloum, N.; Fuhrmann, G.; Schini-Kerth, V.B.; Bronner, C. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J. Exp. Clin. Cancer Res. 2011, CR 30, 41. [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Zhao, J.; Feng, P.; Xing, J.; Wu, H.; Li, Y. Epigenetic mechanism and target therapy of UHRF1 protein complex in malignancies. OncoTargets Ther. 2019, 12, 549–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, O.; Omran, Z.; Hosawi, S.; Hamiche, A.; Bronner, C.; Alhosin, M. Thymoquinone Is a Multitarget Single Epidrug That Inhibits the UHRF1 Protein Complex. Genes 2021, 12, 622. [Google Scholar] [CrossRef] [PubMed]
- Unoki, M. Current and potential anticancer drugs targeting members of the UHRF1 complex including epigenetic modifiers. Recent Pat. Anti-Cancer Drug Discov. 2011, 6, 116–130. [Google Scholar] [CrossRef]
- Ahmad, T.; Ashraf, W.; Ibrahim, A.; Zaayter, L.; Muller, C.D.; Hamiche, A.; Mély, Y.; Bronner, C.; Mousli, M. TIP60 governs the auto-ubiquitination of UHRF1 through USP7 dissociation from the UHRF1/USP7 complex. Int. J. Oncol. 2021, 59, 89. [Google Scholar] [CrossRef]
- Choudhry, H.; Zamzami, M.A.; Omran, Z.; Wu, W.; Mousli, M.; Bronner, C.; Alhosin, M. Targeting microRNA/UHRF1 pathways as a novel strategy for cancer therapy. Oncol. Lett. 2018, 15, 3–10. [Google Scholar] [CrossRef]
- Felle, M.; Joppien, S.; Németh, A.; Diermeier, S.; Thalhammer, V.; Dobner, T.; Kremmer, E.; Kappler, R.; Längst, G. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011, 39, 8355–8365. [Google Scholar] [CrossRef] [Green Version]
- Bronner, C.; Alhosin, M.; Hamiche, A.; Mousli, M. Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes 2019, 10, 65. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [Green Version]
- Ren, W.; Fan, H.; Grimm, S.A.; Guo, Y.; Kim, J.J.; Yin, J.; Li, L.; Petell, C.J.; Tan, X.F.; Zhang, Z.M.; et al. Direct readout of heterochromatic H3K9me3 regulates DNMT1-mediated maintenance DNA methylation. Proc. Natl. Acad. Sci. USA 2020, 117, 18439–18447. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Bronner, C.; Krifa, M.; Mousli, M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem. Pharmacol. 2013, 86, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Kilin, V.; Gavvala, K.; Barthes, N.P.; Michel, B.Y.; Shin, D.; Boudier, C.; Mauffret, O.; Yashchuk, V.; Mousli, M.; Ruff, M.; et al. Dynamics of Methylated Cytosine Flipping by UHRF1. J. Am. Chem. Soc. 2017, 139, 2520–2528. [Google Scholar] [CrossRef] [Green Version]
- Bronner, C.; Fuhrmann, G.; Chédin, F.L.; Macaluso, M.; Dhe-Paganon, S. UHRF1 Links the Histone code and DNA Methylation to ensure Faithful Epigenetic Memory Inheritance. Genet. Epigenet. 2010, 2009, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Wang, L.; Du, Y.; Xie, S.; Yang, X.; Lian, F.; Zhou, Z.; Qian, C. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 2018, 46, 3218–3231. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gao, Q.; Li, P.; Zhao, Q.; Zhang, J.; Li, J.; Koseki, H.; Wong, J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 2013, 4, 1563. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Chen, H.; Guo, X.; Wang, Z.; Sowa, M.E.; Zheng, L.; Hu, S.; Zeng, P.; Guo, R.; Diao, J.; et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc. Natl. Acad. Sci. USA 2012, 109, 4828–4833. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Yang, H.; Fang, J.; Ma, L.; Gong, R.; Wang, P.; Li, Z.; Xu, Y. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat. Commun. 2015, 6, 7023. [Google Scholar] [CrossRef]
- Dar, A.; Shibata, E.; Dutta, A. Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Mol. Cell. Biol. 2013, 33, 3309–3320. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.T.; Kuo, Y.C.; Hung, J.J.; Huang, C.H.; Chen, W.Y.; Chou, T.Y.; Chen, Y.; Chen, Y.J.; Chen, Y.J.; Cheng, W.C.; et al. K63-polyubiquitinated HAUSP deubiquitinates HIF-1α and dictates H3K56 acetylation promoting hypoxia-induced tumour progression. Nat. Commun. 2016, 7, 13644. [Google Scholar] [CrossRef] [Green Version]
- Gagarina, V.; Bojagora, A.; Lacdao, I.K.; Luthra, N.; Pfoh, R.; Mohseni, S.; Chaharlangi, D.; Tan, N.; Saridakis, V. Structural Basis of the Interaction Between Ubiquitin Specific Protease 7 and Enhancer of Zeste Homolog 2. J. Mol. Biol. 2020, 432, 897–912. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jiang, W.; Zhou, P.; Liu, L.; Wan, X.; Yuan, X.; Wang, X.; Chen, M.; Chen, J.; Yang, J.; et al. Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PLoS ONE 2015, 10, e0145023. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Cui, Y.; Xu, Q.; Jiang, Y. Stabilization of LSD1 by deubiquitinating enzyme USP7 promotes glioblastoma cell tumorigenesis and metastasis through suppression of the p53 signaling pathway. Oncol. Rep. 2016, 36, 2935–2945. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Feng, J.; Li, L.; Lin, L.; Ji, J.; Lin, C.; Liu, L.; Zhang, N.; Duan, D.; Li, Z.; et al. Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 2020, 21, e48597. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ma, S.; Song, N.; Li, X.; Liu, L.; Yang, S.; Ding, X.; Shan, L.; Zhou, X.; Su, D.; et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J. Clin. Investig. 2016, 126, 2205–2220. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.M.; Rothbart, S.B.; Allison, D.F.; Cai, Q.; Harrison, J.S.; Li, L.; Wang, Y.; Strahl, B.D.; Wang, G.G.; Song, J. An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell Rep. 2015, 12, 1400–1406. [Google Scholar] [CrossRef] [Green Version]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and function of mammalian DNA methyltransferases. ChemBioChem A Eur. J. Chem. Biol. 2011, 12, 206–222. [Google Scholar] [CrossRef]
- Walton, E.L.; Francastel, C.; Velasco, G. Maintenance of DNA methylation: Dnmt3b joins the dance. Epigenetics 2011, 6, 1373–1377. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.K. DNMT1, A key drug target in triple-negative breast cancer. Semin. Cancer Biol. 2021, 72, 198–213. [Google Scholar] [CrossRef]
- Wong, K.K. DNMT1 as a therapeutic target in pancreatic cancer: Mechanisms and clinical implications. Cell. Oncol. 2020, 43, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Hirohashi, S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis 2007, 28, 2434–2442. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Song, J.; Wang, Y.; Zhao, Y.; Guda, K.; Yang, S.; Kao, H.Y.; Xu, Y.; Willis, J.; Markowitz, S.D.; et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 2010, 3, ra80. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Leonhardt, H.; Spada, F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 2011, 112, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, Y.; Markovtsov, V.; Lang, W.; Sharma, P.; Pearsall, D.; Warner, J.; Franci, C.; Huang, B.; Huang, J.; Yam, G.C.; et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell 2005, 16, 5621–5629. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Alhosin, M.; Papin, C.; Ouararhni, K.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mély, Y.; Hamiche, A.; et al. Thymoquinone challenges UHRF1 to commit auto-ubiquitination: A key event for apoptosis induction in cancer cells. Oncotarget 2018, 9, 28599–28611. [Google Scholar] [CrossRef]
- Hashimoto, H.; Horton, J.R.; Zhang, X.; Bostick, M.; Jacobsen, S.E.; Cheng, X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008, 455, 826–829. [Google Scholar] [CrossRef]
- Avvakumov, G.V.; Walker, J.R.; Xue, S.; Li, Y.; Duan, S.; Bronner, C.; Arrowsmith, C.H.; Dhe-Paganon, S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008, 455, 822–825. [Google Scholar] [CrossRef]
- Bostick, M.; Kim, J.K.; Estève, P.O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef] [Green Version]
- Sharif, J.; Muto, M.; Takebayashi, S.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef]
- Kim, J.K.; Estève, P.O.; Jacobsen, S.E.; Pradhan, S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009, 37, 493–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achour, M.; Fuhrmann, G.; Alhosin, M.; Rondé, P.; Chataigneau, T.; Mousli, M.; Schini-Kerth, V.B.; Bronner, C. UHRF1 recruits the histone acetyltransferase Tip60 and controls its expression and activity. Biochem. Biophys. Res. Commun. 2009, 390, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, W.; Bronner, C.; Zaayter, L.; Ahmad, T.; Richert, L.; Alhosin, M.; Ibrahim, A.; Hamiche, A.; Mely, Y.; Mousli, M. Interaction of the epigenetic integrator UHRF1 with the MYST domain of TIP60 inside the cell. J. Exp. Clin. Cancer Res. 2017, CR36, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unoki, M.; Nishidate, T.; Nakamura, Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 2004, 23, 7601–7610. [Google Scholar] [CrossRef] [Green Version]
- Bronner, C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci. Signal. 2011, 4, pe3. [Google Scholar] [CrossRef]
- Harrison, J.S.; Cornett, E.M.; Goldfarb, D.; DaRosa, P.A.; Li, Z.M.; Yan, F.; Dickson, B.M.; Guo, A.H.; Cantu, D.V.; Kaustov, L.; et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife 2016, 5, e17101. [Google Scholar] [CrossRef] [Green Version]
- Houliston, R.S.; Lemak, A.; Iqbal, A.; Ivanochko, D.; Duan, S.; Kaustov, L.; Ong, M.S.; Fan, L.; Senisterra, G.; Brown, P.J.; et al. Conformational dynamics of the TTD-PHD histone reader module of the UHRF1 epigenetic regulator reveals multiple histone-binding states, allosteric regulation, and druggability. J. Biol. Chem. 2017, 292, 20947–20959. [Google Scholar] [CrossRef] [Green Version]
- Alhosin, M.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mousli, M.; Bronner, C. Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 2016, CR35, 174. [Google Scholar] [CrossRef] [Green Version]
- Beck, A.; Trippel, F.; Wagner, A.; Joppien, S.; Felle, M.; Vokuhl, C.; Schwarzmayr, T.; Strom, T.M.; von Schweinitz, D.; Längst, G.; et al. Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma. Clin. Epigenet. 2018, 10, 27. [Google Scholar] [CrossRef] [Green Version]
- Sambuudash, O.; Kim, H.S.; Cho, M.Y. Lack of Aberrant Methylation in an Adjacent Area of Left-Sided Colorectal Cancer. Yonsei Med. J. 2017, 58, 749–755. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, A.; Yamaguchi, L.; Sharif, J.; Johmura, Y.; Kawamura, T.; Nakanishi, K.; Shimamura, S.; Arita, K.; Kodama, T.; Ishikawa, F.; et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 2013, 502, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Misaki, T.; Yamaguchi, L.; Sun, J.; Orii, M.; Nishiyama, A.; Nakanishi, M. The replication foci targeting sequence (RFTS) of DNMT1 functions as a potent histone H3 binding domain regulated by autoinhibition. Biochem. Biophys. Res. Commun. 2016, 470, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Wolf, P.; Liu, N.; Link, S.; Smets, M.; La Mastra, F.; Forné, I.; Pichler, G.; Hörl, D.; Fellinger, K.; et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015, 25, 911–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, D.J.; Ma, Y.; Dickson, K.A. Histone Monoubiquitination in Chromatin Remodelling: Focus on the Histone H2B Interactome and Cancer. Cancers 2020, 12, 3462. [Google Scholar] [CrossRef]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Vaissière, T.; Sawan, C.; Herceg, Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 2008, 659, 40–48. [Google Scholar] [CrossRef]
- Yan, Q.; Chen, B.J.; Hu, S.; Qi, S.L.; Li, L.Y.; Yang, J.F.; Zhou, H.; Yang, C.C.; Chen, L.J.; Du, J. Emerging role of RNF2 in cancer: From bench to bedside. J. Cell. Physiol. 2021, 236, 5453–5465. [Google Scholar] [CrossRef]
- Marsh, D.J.; Dickson, K.A. Writing Histone Monoubiquitination in Human Malignancy—The Role of RING Finger E3 Ubiquitin Ligases. Genes 2019, 10, 67. [Google Scholar] [CrossRef] [Green Version]
- Cohen, I.; Bar, C.; Ezhkova, E. Activity of PRC1 and Histone H2AK119 Monoubiquitination: Revising Popular Misconceptions. BioEssays News Rev. Mol. Cell. Dev. Biol. 2020, 42, e1900192. [Google Scholar] [CrossRef]
- Benitz, S.; Regel, I.; Reinhard, T.; Popp, A.; Schäffer, I.; Raulefs, S.; Kong, B.; Esposito, I.; Michalski, C.W.; Kleeff, J. Polycomb repressor complex 1 promotes gene silencing through H2AK119 mono-ubiquitination in acinar-to-ductal metaplasia and pancreatic cancer cells. Oncotarget 2016, 7, 11424–11433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkari, F.; Wheaton, K.; La Delfa, A.; Mohamed, M.; Shaikh, F.; Khatun, R.; Arrowsmith, C.H.; Frappier, L.; Saridakis, V.; Sheng, Y. Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1. J. Biol. Chem. 2013, 288, 16975–16985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chan, H.L.; Garcia-Martinez, L.; Karl, D.L.; Weich, N.; Slingerland, J.M.; Verdun, R.E.; Morey, L. Estrogen induces dynamic ERα and RING1B recruitment to control gene and enhancer activities in luminal breast cancer. Sci. Adv. 2020, 6, eaaz7249. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.L.; Beckedorff, F.; Zhang, Y.; Garcia-Huidobro, J.; Jiang, H.; Colaprico, A.; Bilbao, D.; Figueroa, M.E.; LaCava, J.; Shiekhattar, R.; et al. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 2018, 9, 3377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benitz, S.; Straub, T.; Mahajan, U.M.; Mutter, J.; Czemmel, S.; Unruh, T.; Wingerath, B.; Deubler, S.; Fahr, L.; Cheng, T.; et al. Ring1b-dependent epigenetic remodelling is an essential prerequisite for pancreatic carcinogenesis. Gut 2019, 68, 2007–2018. [Google Scholar] [CrossRef]
- Sánchez-Molina, S.; Figuerola-Bou, E.; Blanco, E.; Sánchez-Jiménez, M.; Táboas, P.; Gómez, S.; Ballaré, C.; García-Domínguez, D.J.; Prada, E.; Hontecillas-Prieto, L.; et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci. Adv. 2020, 6, eaba3058. [Google Scholar] [CrossRef]
- de Bie, P.; Zaaroor-Regev, D.; Ciechanover, A. Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochem. Biophys. Res. Commun. 2010, 400, 389–395. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Wheaton, K.; Sarkari, F.; Johns, B.S.; Davarinejad, H.; Egorova, O.; Kaustov, L.; Raught, B.; Saridakis, V.; Sheng, Y. UbE2E1/UBCH6 Is a Critical In Vivo E2 for the PRC1-catalyzed Ubiquitination of H2A at Lys-119. J. Biol. Chem. 2017, 292, 2893–2902. [Google Scholar] [CrossRef] [Green Version]
- van der Knaap, J.A.; Kumar, B.R.; Moshkin, Y.M.; Langenberg, K.; Krijgsveld, J.; Heck, A.J.; Karch, F.; Verrijzer, C.P. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 2005, 17, 695–707. [Google Scholar] [CrossRef]
- Squatrito, M.; Gorrini, C.; Amati, B. Tip60 in DNA damage response and growth control: Many tricks in one HAT. Trends Cell Biol. 2006, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Judes, G.; Rifaï, K.; Ngollo, M.; Daures, M.; Bignon, Y.J.; Penault-Llorca, F.; Bernard-Gallon, D. A bivalent role of TIP60 histone acetyl transferase in human cancer. Epigenomics 2015, 7, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Judes, G.; Dubois, L.; Rifaï, K.; Idrissou, M.; Mishellany, F.; Pajon, A.; Besse, S.; Daures, M.; Degoul, F.; Bignon, Y.J.; et al. TIP60, an actor in acetylation of H3K4 and tumor development in breast cancer. Epigenomics 2018, 10, 1415–1430. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Luo, J.; Zhang, W.; Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 2006, 24, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Sykes, S.M.; Mellert, H.S.; Holbert, M.A.; Li, K.; Marmorstein, R.; Lane, W.S.; McMahon, S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 2006, 24, 841–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altun, M.; Kramer, H.B.; Willems, L.I.; McDermott, J.L.; Leach, C.A.; Goldenberg, S.J.; Kumar, K.G.; Konietzny, R.; Fischer, R.; Kogan, E.; et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011, 18, 1401–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Song, C.; Young, N.L.; Sperling, A.S.; Xu, F.; Sridharan, R.; Conway, A.E.; Garcia, B.A.; Plath, K.; Clark, A.T.; et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol. Cell 2009, 33, 417–427. [Google Scholar] [CrossRef] [Green Version]
- Stejskal, S.; Stepka, K.; Tesarova, L.; Stejskal, K.; Matejkova, M.; Simara, P.; Zdrahal, Z.; Koutna, I. Cell cycle-dependent changes in H3K56ac in human cells. Cell Cycle 2015, 14, 3851–3863. [Google Scholar] [CrossRef]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar] [CrossRef]
- Yamagishi, M.; Uchimaru, K. Targeting EZH2 in cancer therapy. Curr. Opin. Oncol. 2017, 29, 375–381. [Google Scholar] [CrossRef]
- Lim, S.; Janzer, A.; Becker, A.; Zimmer, A.; Schüle, R.; Buettner, R.; Kirfel, J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010, 31, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Nottke, A.C.; Shi, Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008, 20, 316–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukada, Y.; Ishitani, T.; Nakayama, K.I. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010, 24, 432–437. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Yonezawa, M.; Ye, J.; Jenuwein, T.; Grummt, I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 2010, 17, 445–450. [Google Scholar] [CrossRef]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2, a novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Kirmizis, A.; Bartley, S.M.; Kuzmichev, A.; Margueron, R.; Reinberg, D.; Green, R.; Farnham, P.J. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004, 18, 1592–1605. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.I.; Jenner, R.G.; Boyer, L.A.; Guenther, M.G.; Levine, S.S.; Kumar, R.M.; Chevalier, B.; Johnstone, S.E.; Cole, M.F.; Isono, K.; et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006, 125, 301–313. [Google Scholar] [CrossRef] [Green Version]
- Mas, Y.M.S.; Barbon, M.; Teyssier, C.; Déméné, H.; Carvalho, J.E.; Bird, L.E.; Lebedev, A.; Fattori, J.; Schubert, M.; Dumas, C.; et al. The Human Mixed Lineage Leukemia 5 (MLL5), a Sequentially and Structurally Divergent SET Domain-Containing Protein with No Intrinsic Catalytic Activity. PLoS ONE 2016, 11, e0165139. [Google Scholar]
- Zhou, P.; Wang, Z.; Yuan, X.; Zhou, C.; Liu, L.; Wan, X.; Zhang, F.; Ding, X.; Wang, C.; Xiong, S.; et al. Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1). J. Biol. Chem. 2013, 288, 17532–17543. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, J.; Kahl, P.; Mertens, C.; Rogenhofer, S.; Hauser, S.; Hartmann, W.; Bastian, P.J.; Büttner, R.; Müller, S.C.; von Ruecker, A. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int. J. Cancer 2010, 127, 2360–2366. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Xu, J.; Zhang, J.; Xie, D.; Ye, H.; Xiao, Z.; Cai, M.; Xu, K.; Zeng, Y.; Li, H.; et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum. Pathol. 2012, 43, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Benard, A.; Goossens-Beumer, I.J.; van Hoesel, A.Q.; de Graaf, W.; Horati, H.; Putter, H.; Zeestraten, E.C.; van de Velde, C.J.; Kuppen, P.J. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer 2014, 14, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prusevich, P.; Kalin, J.H.; Ming, S.A.; Basso, M.; Givens, J.; Li, X.; Hu, J.; Taylor, M.S.; Cieniewicz, A.M.; Hsiao, P.Y.; et al. A selective phenelzine analogue inhibitor of histone demethylase LSD1. ACS Chem. Biol. 2014, 9, 1284–1293. [Google Scholar] [CrossRef]
- Amente, S.; Milazzo, G.; Sorrentino, M.C.; Ambrosio, S.; Di Palo, G.; Lania, L.; Perini, G.; Majello, B. Lysine-specific demethylase (LSD1/KDM1A) and MYCN cooperatively repress tumor suppressor genes in neuroblastoma. Oncotarget 2015, 6, 14572–14583. [Google Scholar] [CrossRef] [Green Version]
- Lv, T.; Yuan, D.; Miao, X.; Lv, Y.; Zhan, P.; Shen, X.; Song, Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE 2012, 7, e35065. [Google Scholar]
- Ketscher, A.; Jilg, C.A.; Willmann, D.; Hummel, B.; Imhof, A.; Rüsseler, V.; Hölz, S.; Metzger, E.; Müller, J.M.; Schüle, R. LSD1 controls metastasis of androgen-independent prostate cancer cells through PXN and LPAR6. Oncogenesis 2014, 3, e120. [Google Scholar] [CrossRef]
- Shao, P.; Liu, Q.; Maina, P.K.; Cui, J.; Bair, T.B.; Li, T.; Umesalma, S.; Zhang, W.; Qi, H.H. Histone demethylase PHF8 promotes epithelial to mesenchymal transition and breast tumorigenesis. Nucleic Acids Res. 2017, 45, 1687–1702. [Google Scholar] [CrossRef]
- Liu, Q.; Pang, J.; Wang, L.A.; Huang, Z.; Xu, J.; Yang, X.; Xie, Q.; Huang, Y.; Tang, T.; Tong, D.; et al. Histone demethylase PHF8 drives neuroendocrine prostate cancer progression by epigenetically upregulating FOXA2. J. Pathol. 2021, 253, 106–118. [Google Scholar] [CrossRef]
- Zhu, G.; Liu, L.; She, L.; Tan, H.; Wei, M.; Chen, C.; Su, Z.; Huang, D.; Tian, Y.; Qiu, Y.; et al. Elevated expression of histone demethylase PHF8 associates with adverse prognosis in patients of laryngeal and hypopharyngeal squamous cell carcinoma. Epigenomics 2015, 7, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, Y.; Fang, J.; Xiao, J.; Zhu, T.; Chen, F.; Wang, P.; Li, Z.; Yang, H.; Xu, Y. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 2013, 288, 1329–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 2014, 54, 905–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kategaya, L.; di Lello, P.; Rougé, L.; Pastor, R.; Clark, K.R.; Drummond, J.; Kleinheinz, T.; Lin, E.; Upton, J.P.; Prakash, S.; et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2009, 550, 534–538. [Google Scholar] [CrossRef] [Green Version]
- Pozhidaeva, A.; Valles, G.; Wang, F.; Wu, J.; Sterner, D.E.; Nguyen, P.; Weinstock, J.; Kumar, K.G.S.; Kanyo, J.; Wright, D.; et al. USP7-Specific Inhibitors Target and Modify the Enzyme’s Active Site via Distinct Chemical Mechanisms. Cell Chem. Biol. 2017, 24, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Nininahazwe, L.; Liu, B.; He, C.; Zhang, H.; Chen, Z.S. The emerging nature of Ubiquitin-specific protease 2017, 7 (USP7): A new target in cancer therapy. Drug Discov. Today 2021, 26, 490–502. [Google Scholar] [CrossRef] [PubMed]

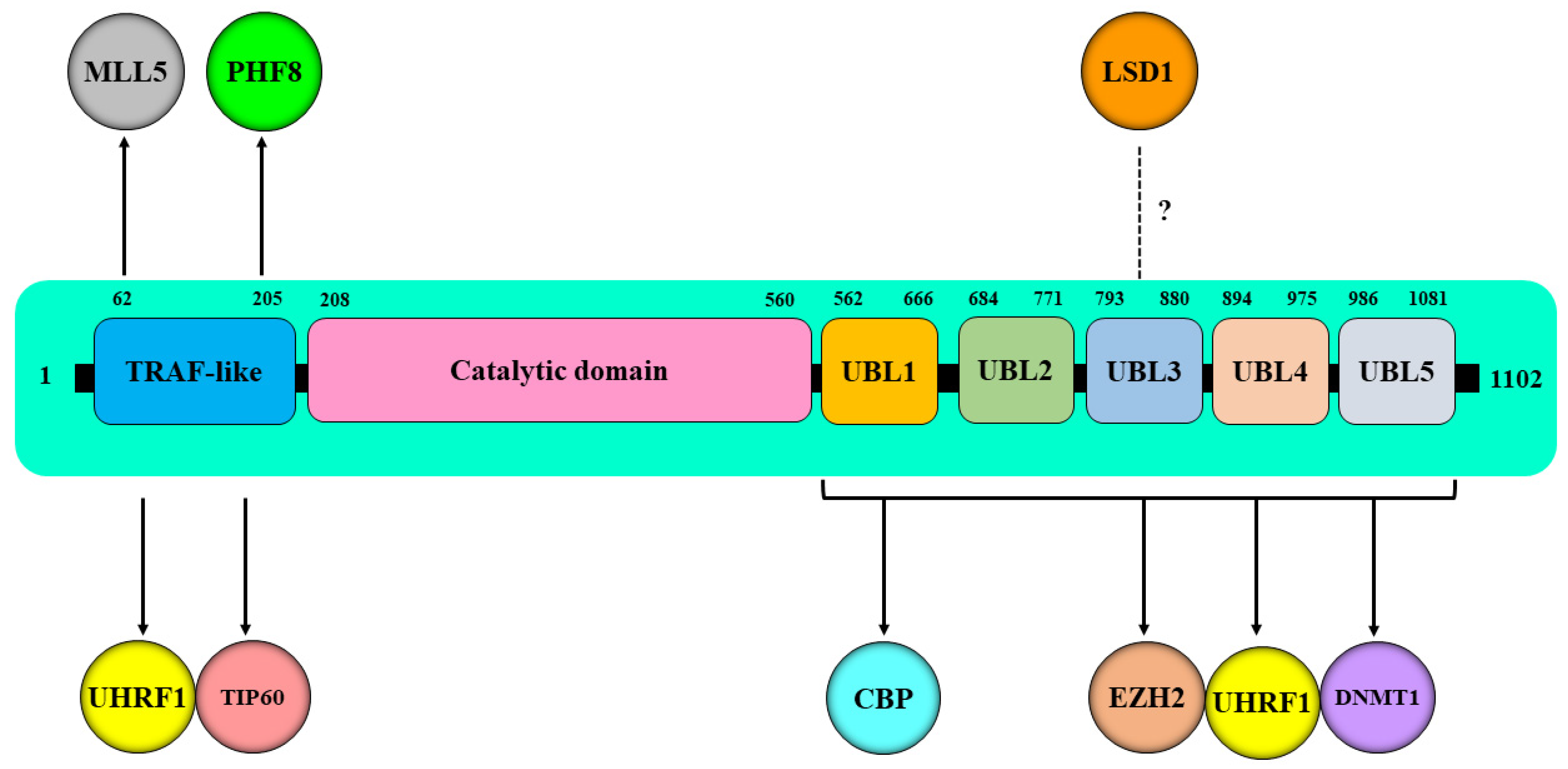

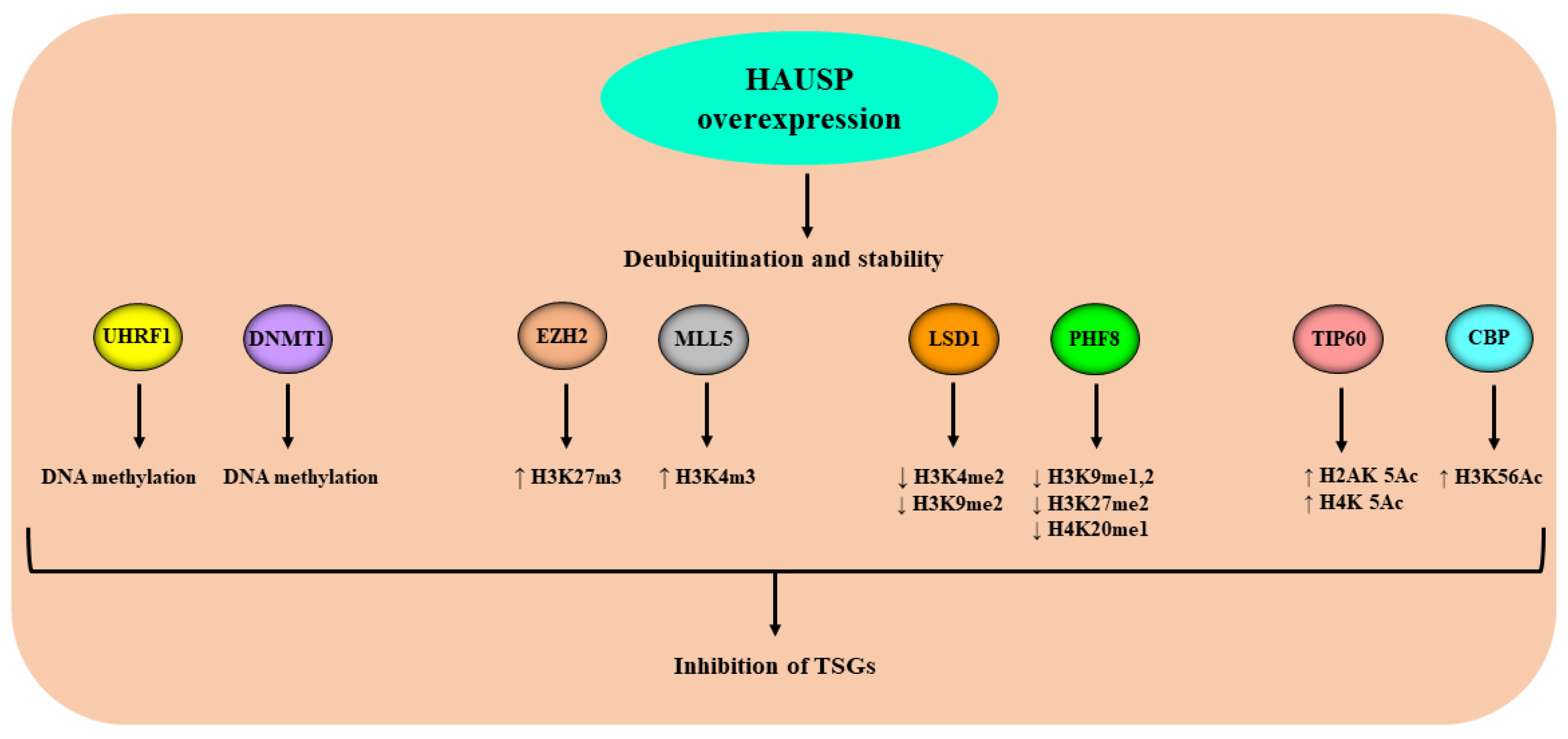
| Epi-Partner | Role of Epi-Partner | HAUSP Interaction Site | Epi-Partner Interaction Site | Related Epigenetic Events | Refs. |
|---|---|---|---|---|---|
| UHRF1 | Reader of both epigenetic marks (DNA methylation and histone code) | UBL1 domain (residues 560–664) | A linker region encompassing amino acids 600–687 between the SRA and RING finger domains of UHRF1 | Promoting the stability of UHRF1 through HAUSP-dependent deubiquitination | [48] |
| TRAF-like domain | SRA domain | Promoting the stability of UHRF1 through HAUSP-dependent deubiquitination | [38] | ||
| (UBL1-2) domains | A polybasic region (PBR) in the C terminus | Promoting the association of the TTD-PHD domains of UHRF1 with chromatin and, hence, efficient H3K9me3 binding | [57] | ||
| DNMT1 | DNA methyltransferase 1 | C-terminal domain | Targeting sequence (TS) domain | Promoting the stability of DNMT1 through HAUSP-dependent deubiquitination | [38] |
| UBL domains (residues 560–1102) | KG linker (residues 1109–1119) | Promoting the stability of DNMT1 through acetylation of KG linker of DNMT1 | [49] | ||
| TIP60 | Histone acetyltransferase | TRAF-like domain | Increased levels of H2AK 5 and H4K5 | [50] | |
| CBP | Histone acetyltransferase | Region encompassing amino acids 600–687 | CH3 domain | Increased levels of H3K56Ac | [51] |
| MLL5 | Histone lysine methyltransferase | TRAF domain | Multiple domains | Increased levels of H3K4m3 | [53] |
| EZH2 | Histone lysine N-methyltransferase | C-terminal region | 489PRKKKRK495 region | Increased levels of H3K27m3 | [52] |
| LSD1 | Lysine specific demethylase 1 | [54] | |||
| A region encompassing amino acids 600–687 | Demethylation of H3K4me2 and H3K9me2 | [55] | |||
| PHF8 | Histone lysine demethylase | TRAF-like domain | The C-terminal region | Demethylation of H3K9me1,2, H3K27me2, and H4K20me1 | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, O.; Alhosin, M. HAUSP Is a Key Epigenetic Regulator of the Chromatin Effector Proteins. Genes 2022, 13, 42. https://doi.org/10.3390/genes13010042
Abdullah O, Alhosin M. HAUSP Is a Key Epigenetic Regulator of the Chromatin Effector Proteins. Genes. 2022; 13(1):42. https://doi.org/10.3390/genes13010042
Chicago/Turabian StyleAbdullah, Omeima, and Mahmoud Alhosin. 2022. "HAUSP Is a Key Epigenetic Regulator of the Chromatin Effector Proteins" Genes 13, no. 1: 42. https://doi.org/10.3390/genes13010042
APA StyleAbdullah, O., & Alhosin, M. (2022). HAUSP Is a Key Epigenetic Regulator of the Chromatin Effector Proteins. Genes, 13(1), 42. https://doi.org/10.3390/genes13010042






