Rapid Cis–Trans Coevolution Driven by a Novel Gene Retroposed from a Eukaryotic Conserved CCR4–NOT Component in Drosophila
Abstract
:1. Introduction
2. Materials and Methods
2.1. piggyBac Vector Construction
2.2. Injections and Screening for Transgenics
2.3. Chromatin Immunoprecipitation and Sequencing
2.4. Sequencing and Read Mapping
2.5. Signal and Peak Calling
2.6. Sex-Bias and Spermatogenesis Expression
2.7. DREME
2.8. Motif Analysis
2.9. Exon Bias of Motifs
2.10. Promoter Motif Frequency Analysis
2.11. Statistical Analysis
2.12. Population Genetics
2.13. CRISPR-Cas9 Mediated Zeus Deletion
2.14. RNA Extraction, Library Preparation and Differential Expression Genes Analysis
3. Results
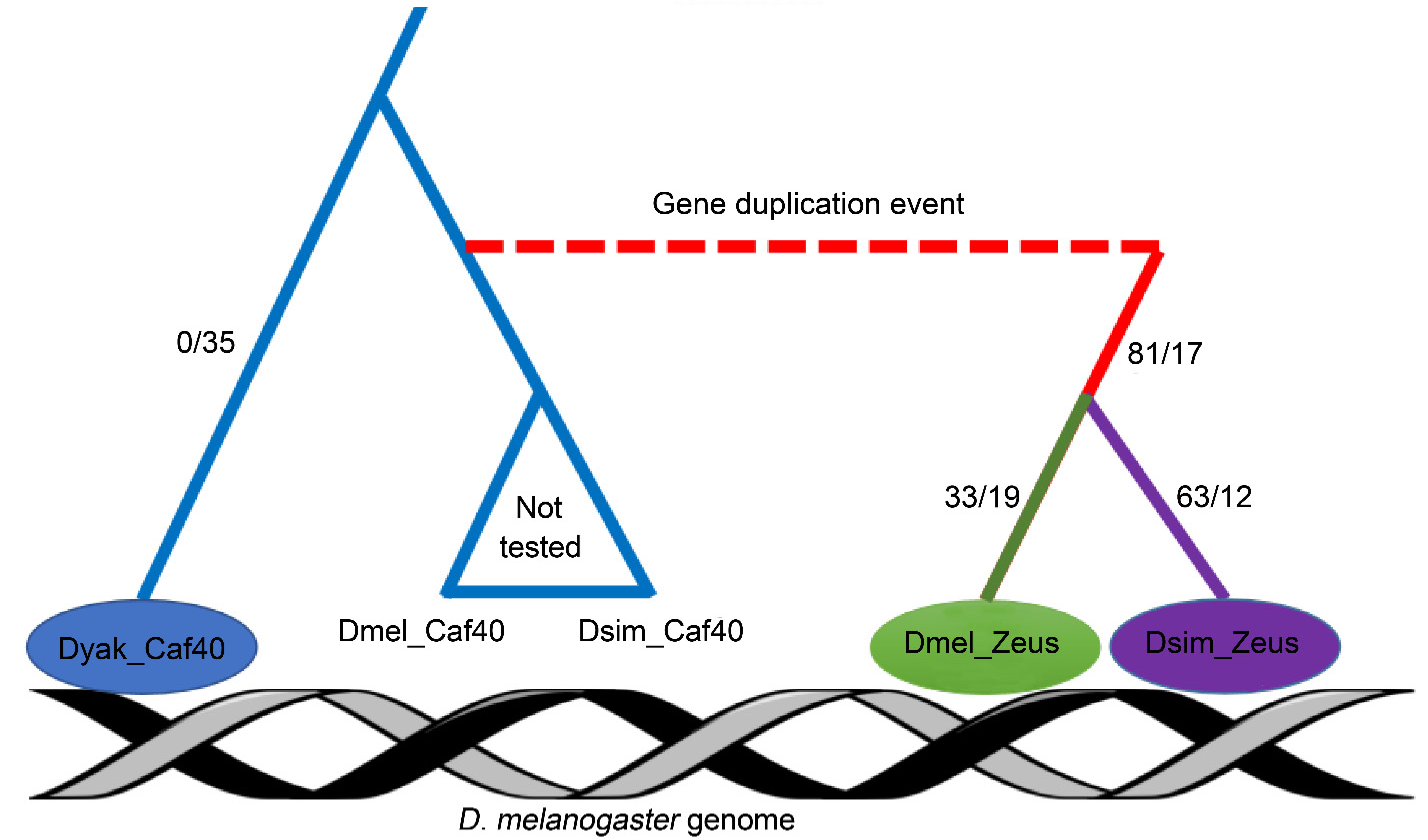
3.1. Divergent ChIP-Seq Profile between D. melanogaster Zeus, D. simulans Zeus and D. yakuba Caf40
3.2. Zeus Gained Affinity for Sex-Biased Genes on Both X Chromosome and Chromosome 4
3.3. Zeus-Derived Genome-Wide Frequency of CAZAM-Motif Variation and Motif Redistribution between Drosophila Species with and without Zeus Gene
3.4. Origination of Zeus Reshaped Selection Pressure Variance of the Motif across Species
3.5. Zeus-Regulated Gene Expression Does Not Depend on CAZAM Binding in the Whole Testes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.D.; Krinsky, B.H.; Long, M.Y. New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 2013, 14, 645–660. [Google Scholar] [CrossRef] [Green Version]
- Long, M.Y.; VanKuren, N.W.; Chen, S.D.; Vibranovski, M.D. New gene evolution: Little did we know. Annu. Rev. Genet. 2013, 47, 307–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, M.; Betran, E.; Thornton, K.; Wang, W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003, 4, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ernst, J.; Kharchenko, P.V.; Kheradpour, P.; Negre, N.; Eaton, M.L.; Landolin, J.M.; Bristow, C.A.; Ma, L.; Lin, M.F.; et al. Identification of functional elements and regulatory circuits by drosophila modencode. Science 2010, 330, 1787–1797. [Google Scholar] [PubMed] [Green Version]
- Lee, Y.C.G.; Ventura, I.M.; Rice, G.R.; Chen, D.-Y.; Colmenares, S.U.; Long, M. Rapid evolution of gained essential developmental functions of a young gene via interactions with other essential genes. Mol. Biol. Evol. 2019, 36, 2212–2226. [Google Scholar] [CrossRef]
- Schmidt, D.; Wilson, M.D.; Ballester, B.; Schwalie, P.C.; Brown, G.D.; Marshall, A.; Kutter, C.; Watt, S.; Martinez-Jimenez, C.P.; Mackay, S.; et al. Five-vertebrate chip-seq reveals the evolutionary dynamics of transcription factor binding. Science 2010, 328, 1036–1040. [Google Scholar] [CrossRef]
- Paris, M.; Kaplan, T.; Li, X.Y.; Villalta, J.E.; Lott, S.E.; Eisen, M.B. Extensive divergence of transcription factor binding in drosophila embryos with highly conserved gene expression. PLoS Genet. 2013, 9, e1003748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefflova, K.; Thybert, D.; Wilson, M.D.; Streeter, I.; Aleksic, J.; Karagianni, P.; Brazma, A.; Adams, D.J.; Talianidis, I.; Marioni, J.C.; et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell 2013, 154, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Ni, X.C.; Zhang, Y.E.; Negre, N.; Chen, S.; Long, M.Y.; White, K.P. Adaptive evolution and the birth of ctcf binding sites in the drosophila genome. PLoS Biol. 2012, 10, e1001420. [Google Scholar]
- Betran, E.; Thornton, K.; Long, M. Retroposed new genes out of the x in drosophila. Genome Res. 2002, 12, 1854–1859. [Google Scholar]
- Dai, H.; Yoshimatsu, T.F.; Long, M. Retrogene movement within-and between-chromosomes in the evolution of drosophila genomes. Gene 2006, 385, 96–102. [Google Scholar] [CrossRef]
- Emerson, J.; Kaessmann, H.; Betrán, E.; Long, M. Extensive gene traffic on the mammalian x chromosome. Science 2004, 303, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Ventura, I.M.; Blaha, A.; Sgromo, A.; Han, S.; Izaurralde, E.; Long, M. Rapid gene evolution in an ancient post-transcriptional and translational regulatory system compensates for meiotic x chromosomal inactivation. Mol. Biol. Evol. 2021. [Google Scholar] [CrossRef]
- Bai, Y.S.; Casola, C.; Feschotte, C.; Betran, E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in drosophila. Genome Biol. 2007, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quezada-Diaz, J.E.; Muliyil, T.; Rio, J.; Betran, E. Drcd-1 related: A positively selected spermatogenesis retrogene in drosophila. Genetica 2010, 138, 925–937. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.D.; Ni, X.C.; Krinsky, B.H.; Zhang, Y.E.; Vibranovski, M.D.; White, K.P.; Long, M.Y. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J. 2012, 31, 2798–2809. [Google Scholar] [CrossRef] [Green Version]
- Arthur, R.K.; Ma, L.; Slattery, M.; Spokony, R.F.; Ostapenko, A.; Nègre, N.; White, K.P. Evolution of h3k27me3-marked chromatin is linked to gene expression evolution and to patterns of gene duplication and diversification. Genome Res. 2014, 24, 1115–1124. [Google Scholar] [CrossRef] [Green Version]
- Garces, R.G.; Gillon, W.; Pai, E.F. Atomic model of human rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 2007, 16, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Handler, A.M.; Harrell Ii, R.A., 2nd. Germline transformation of drosophila melanogaster with the piggybac transposon vector. Insect Mol. Biol. 1999, 8, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P.; et al. Chip-seq guidelines and practices of the encode and modencode consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. Characterizing complex structural variation in germline and somatic genomes. Trends Genet. 2012, 28, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreos, R.; Ambrosini, G.; Perier, R.C.; Bucher, P. The eukaryotic promoter database: Expansion of epdnew and new promoter analysis tools. Nucleic Acids Res. 2015, 43, D92–D96. [Google Scholar] [CrossRef]
- Gnad, F.; Parsch, J. Sebida: A database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics 2006, 22, 2577–2579. [Google Scholar] [CrossRef] [Green Version]
- Vibranovski, M.D.; Zhang, Y.; Long, M. General gene movement off the x chromosome in the drosophila genus. Genome Res. 2009, 19, 897–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranz, J.M.; Castillo-Davis, C.I.; Meiklejohn, C.D.; Hartl, D.L. Sex-dependent gene expression and evolution of the drosophila transcriptome. Science 2003, 300, 1742–1745. [Google Scholar] [CrossRef]
- Garland, T., Jr.; Dickerman, A.W.; Janis, C.M.; Jones, J.A. Phylogenetic analysis of covariance by computer simulation. Syst. Biol. 1993, 42, 265–292. [Google Scholar] [CrossRef] [Green Version]
- Revell, L.J. Phytools: An r package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. Bedtools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Subgroup. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Neph, S.; Kuehn, M.S.; Reynolds, A.P.; Haugen, E.; Thurman, R.E.; Johnson, A.K.; Rynes, E.; Maurano, M.T.; Vierstra, J.; Thomas, S.; et al. Bedops: High-performance genomic feature operations. Bioinformatics 2012, 28, 1919–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. Fimo: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohlf, F.J.; Sokal, R.R. Statistical Tables; Macmillan: New York, NY, USA, 1995. [Google Scholar]
- Mackay, T.F.C.; Richards, S.; Stone, E.A.; Barbadilla, A.; Ayroles, J.F.; Zhu, D.H.; Casillas, S.; Han, Y.; Magwire, M.M.; Cridland, J.M.; et al. The drosophila melanogaster genetic reference panel. Nature 2012, 482, 173–178. [Google Scholar] [CrossRef] [Green Version]
- McDonald, J.H.; Kreitman, M. Adaptive protein evolution at the adh locus in drosophila. Nature 1991, 351, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Akashi, H. Codon bias evolution in drosophila. Population genetics of mutation-selection drift. Gene 1997, 205, 269–278. [Google Scholar] [CrossRef]
- Eyre-Walker, A.; Hurst, L.D. The evolution of isochores. Nat. Rev. Genet. 2001, 2, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.; Liu, J.L. Crispr/cas9 mediated genome engineering in drosophila. Methods 2014, 69, 128–136. [Google Scholar] [CrossRef]
- VanKuren, N.W.; Long, M. Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat. Ecol. Evol. 2018, 2, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Ukken, F.P.; Rubinstein, C.D.; Thiede, G.; Donohue, L.K.; Cummings, A.M.; O’Connor-Giles, K.M. Highly specific and efficient crispr/cas9-catalyzed homology-directed repair in drosophila. Genetics 2014, 196, 961–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal rna-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.F.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. Panther version 14: More genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Conrad, T.; Akhtar, A. Dosage compensation in drosophila melanogaster: Epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 2012, 13, 123–134. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex chromosome to an autosome in drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landeen, E.L.; Presgraves, D.C. Evolution: From autosomes to sex chromosomes--and back. Curr. Biol. 2013, 23, R848–R850. [Google Scholar] [CrossRef] [Green Version]
- Meiklejohn, C.D.; Landeen, E.L.; Cook, J.M.; Kingan, S.B.; Presgraves, D.C. Sex chromosome-specific regulation in the drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 2011, 9, e1001126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, R.; Valencia, A. A gene network for navigating the literature. Nat. Genet. 2004, 36, 664. [Google Scholar] [CrossRef] [PubMed]
- Ross, B.D.; Rosin, L.; Thomae, A.W.; Hiatt, M.A.; Vermaak, D.; de la Cruz, A.F.; Imhof, A.; Mellone, B.G.; Malik, H.S. Stepwise evolution of essential centromere function in a drosophila neogene. Science 2013, 340, 1211–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L. Dreme: Motif discovery in transcription factor chip-seq data. Bioinformatics 2011, 27, 1653–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, I.K.; McDonald, J.F. Interelement selection in the regulatory region of the copia retrotransposon. J. Mol. Evol. 1998, 47, 670–676. [Google Scholar] [CrossRef] [PubMed]
- He, B.Z.; Holloway, A.K.; Maerkl, S.J.; Kreitman, M. Does positive selection drive transcription factor binding site turnover? A test with drosophila cis-regulatory modules. PLoS Genet. 2011, 7, e1002053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, M.W. Detecting natural selection on cis-regulatory DNA. Genetica 2007, 129, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Haddrill, P.R.; Bachtrog, D.; Andolfatto, P. Positive and negative selection on noncoding DNA in drosophila simulans. Mol. Biol. Evol. 2008, 25, 1825–1834. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.D.; Begun, D.J. Parallel evolution of chimeric fusion genes. Proc. Natl. Acad. Sci. USA 2005, 102, 11373–11378. [Google Scholar] [CrossRef] [Green Version]
- Ellegren, H.; Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007, 8, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Vibranovski, M.D.; Lopes, H.F.; Karr, T.L.; Long, M. Stage-specific expression profiling of drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009, 5, e1000731. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Rappsilber, J.; Chiang, Y.C.; Russell, P.; Mann, M.; Denis, C.L. Purification and characterization of the 1.0 mda ccr4-not complex identifies two novel components of the complex. J. Mol. Biol. 2001, 314, 683–694. [Google Scholar] [CrossRef]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor binding evokes latent differences in DNA binding specificity between hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazemian, M.; Pham, H.; Wolfe, S.A.; Brodsky, M.H.; Sinha, S. Widespread evidence of cooperative DNA binding by transcription factors in drosophila development. Nucleic Acids Res. 2013, 41, 8237–8252. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Siggers, T.; Duyzend, M.H.; Reddy, J.; Khan, S.; Bulyk, M.L. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol. Syst. Biol. 2011, 7, 555. [Google Scholar] [CrossRef] [PubMed]
- Bawankar, P.; Loh, B.; Wohlbold, L.; Schmidt, S.; Izaurralde, E. Not10 and c2orf29/not11 form a conserved module of the ccr4-not complex that docks onto the not1 n-terminal domain. RNA Biol. 2013, 10, 228–244. [Google Scholar] [CrossRef] [Green Version]
- Collart, M.A.; Panasenko, O.O. The ccr4--not complex. Gene 2012, 492, 42–53. [Google Scholar] [CrossRef]
- Temme, C.; Zhang, L.; Kremmer, E.; Ihling, C.; Chartier, A.; Sinz, A.; Simonelig, M.; Wahle, E. Subunits of the drosophila ccr4-not complex and their roles in mrna deadenylation. RNA 2010, 16, 1356–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgromo, A.; Raisch, T.; Bawankar, P.; Bhandari, D.; Chen, Y.; Kuzuoğlu-Öztürk, D.; Weichenrieder, O.; Izaurralde, E. A caf40-binding motif facilitates recruitment of the ccr4-not complex to mrnas targeted by drosophila roquin. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Sgromo, A.; Raisch, T.; Backhaus, C.; Keskeny, C.; Alva, V.; Weichenrieder, O.; Izaurralde, E. Drosophila bag-of-marbles directly interacts with the caf40 subunit of the ccr4–not complex to elicit repression of mrna targets. RNA 2018, 24, 381–395. [Google Scholar] [CrossRef] [Green Version]
- Andolfatto, P. Controlling type-i error of the mcdonald-kreitman test in genomewide scans for selection on noncoding DNA. Genetics 2008, 180, 1767–1771. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krinsky, B.H.; Arthur, R.K.; Xia, S.; Sosa, D.; Arsala, D.; White, K.P.; Long, M. Rapid Cis–Trans Coevolution Driven by a Novel Gene Retroposed from a Eukaryotic Conserved CCR4–NOT Component in Drosophila. Genes 2022, 13, 57. https://doi.org/10.3390/genes13010057
Krinsky BH, Arthur RK, Xia S, Sosa D, Arsala D, White KP, Long M. Rapid Cis–Trans Coevolution Driven by a Novel Gene Retroposed from a Eukaryotic Conserved CCR4–NOT Component in Drosophila. Genes. 2022; 13(1):57. https://doi.org/10.3390/genes13010057
Chicago/Turabian StyleKrinsky, Benjamin H., Robert K. Arthur, Shengqian Xia, Dylan Sosa, Deanna Arsala, Kevin P. White, and Manyuan Long. 2022. "Rapid Cis–Trans Coevolution Driven by a Novel Gene Retroposed from a Eukaryotic Conserved CCR4–NOT Component in Drosophila" Genes 13, no. 1: 57. https://doi.org/10.3390/genes13010057
APA StyleKrinsky, B. H., Arthur, R. K., Xia, S., Sosa, D., Arsala, D., White, K. P., & Long, M. (2022). Rapid Cis–Trans Coevolution Driven by a Novel Gene Retroposed from a Eukaryotic Conserved CCR4–NOT Component in Drosophila. Genes, 13(1), 57. https://doi.org/10.3390/genes13010057






