Sleep Polygenic Risk Score Is Associated with Cognitive Changes over Time
Abstract
:1. Introduction
2. Methods
3. Statistical Analysis
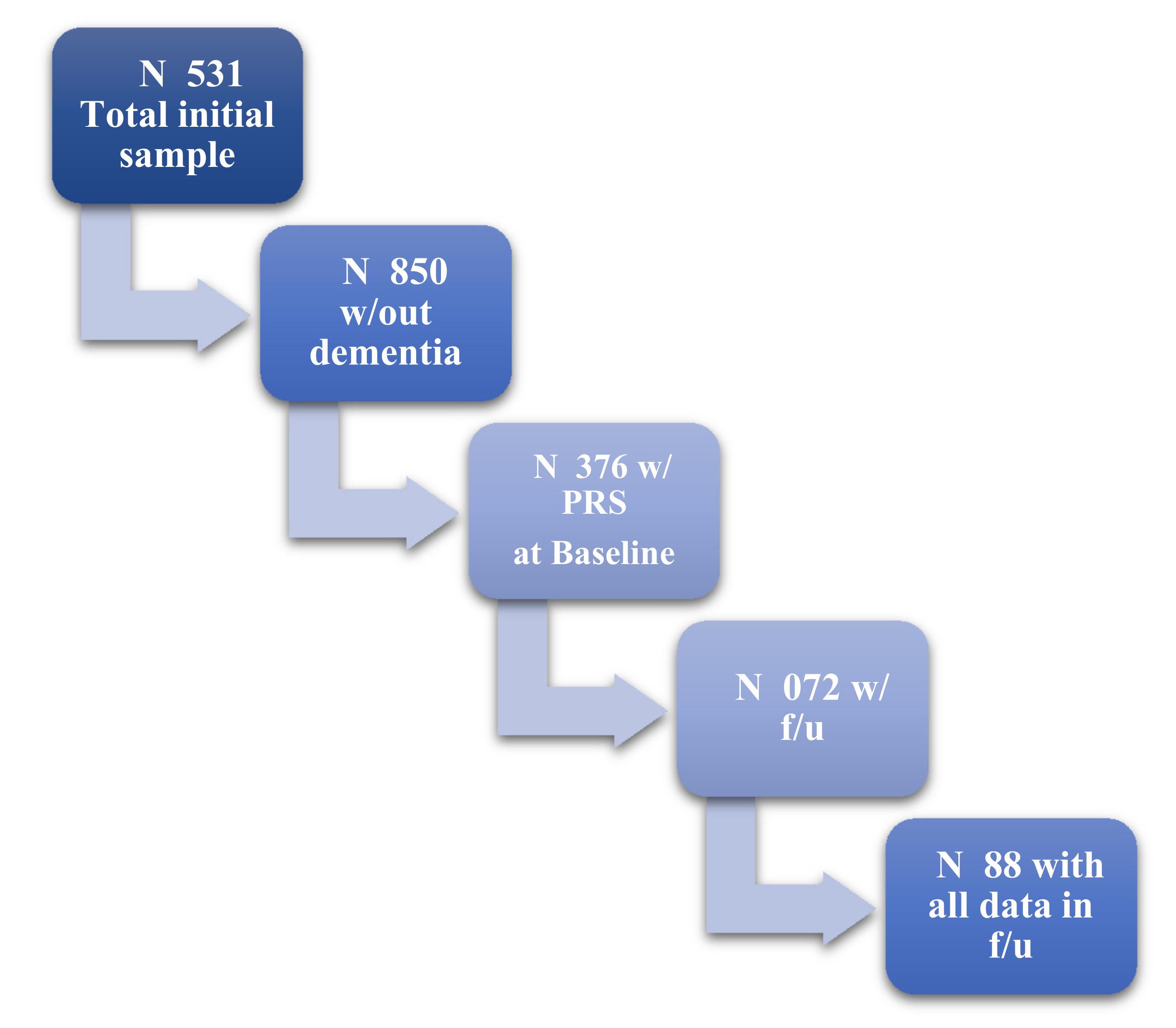
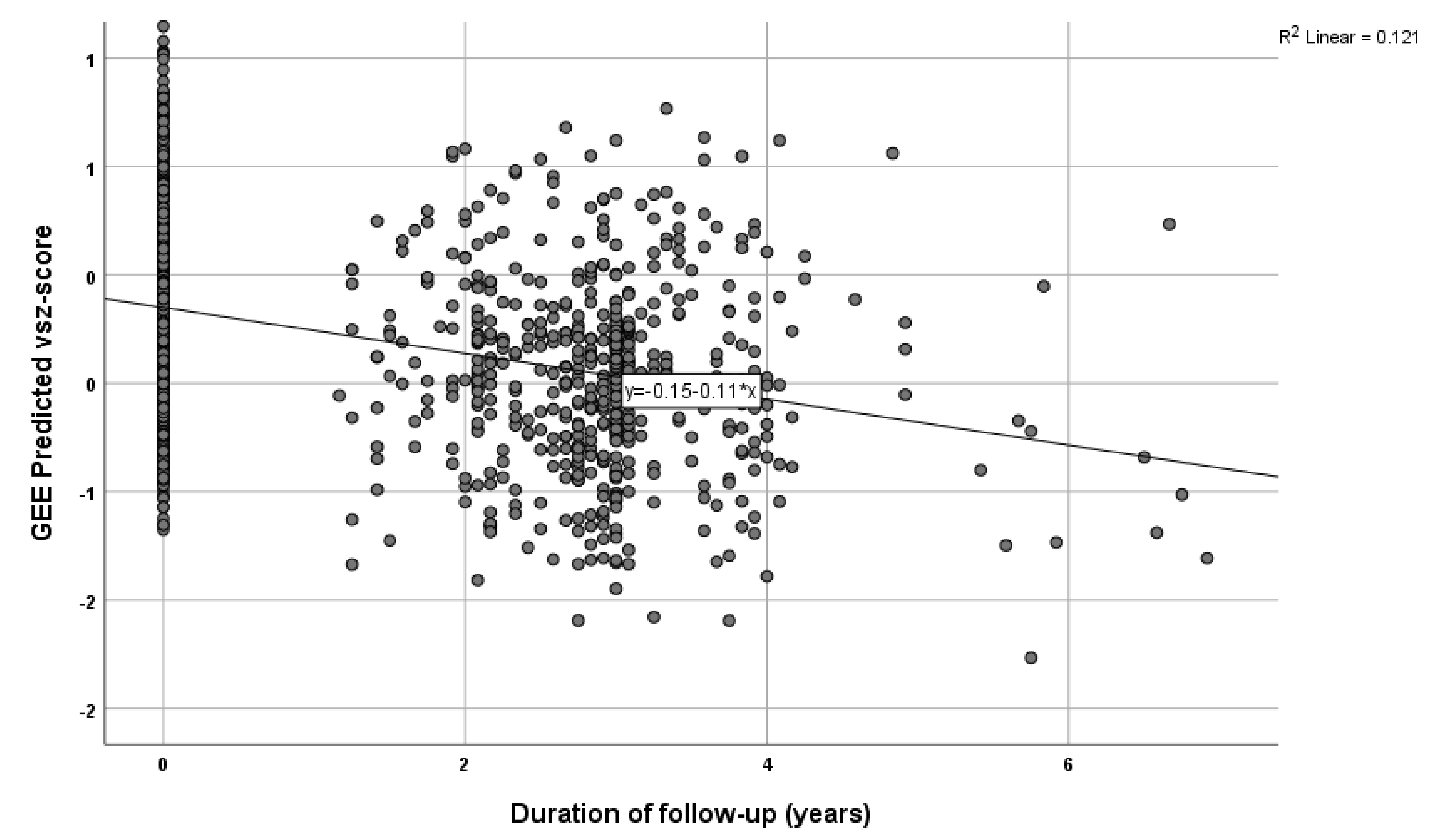
4. Results
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Vitiello, M.V.; Gooneratne, N.S. Sleep in Normal Aging. Sleep Med. Clin. 2018, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tsapanou, A.; Scarmeas, N.; Stern, Y. Sleep and the aging brain. A multifaceted approach. Sleep Sci. 2020, 13, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Tsapanou, A.; Gu, Y.; O’Shea, D.M.; Yannakoulia, M.; Kosmidis, M.; Dardiotis, E.; Hadjigeorgiou, G.; Sakka, P.; Stern, Y.; Scarmeas, N. Sleep quality and duration in relation to memory in the elderly: Initial results from the Hellenic Longitudinal Investigation of Aging and Diet. Neurobiol. Learn. Mem. 2017, 141, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Tsapanou, A.; Gu, Y.; O’Shea, D.M.; Yannakoulia, M.; Kosmidis, M.H.; Dardiotis, E.; Hadjigeorgiou, G.; Sakka, P.; Stern, Y.; Scarmeas, N. Dataset on the associations between sleep quality/duration and cognitive performance in cognitively healthy older adults. Data Brief 2017, 14, 720–723. [Google Scholar] [CrossRef]
- Tsapanou, A.; Gu, Y.; Manly, J.; Schupf, N.; Tang, M.X.; Zimmerman, M.; Scarmeas, N.; Stern, Y. Daytime Sleepiness and Sleep Inadequacy as Risk Factors for Dementia. Dement. Geriatr. Cogn. Dis. Extra 2015, 5, 286–295. [Google Scholar] [CrossRef]
- Tsapanou, A.; Gu, Y.; O’Shea, D.; Eich, T.; Tang, M.X.; Schupf, N.; Manly, J.; Zimmerman, M.; Scarmeas, N.; Stern, Y. Daytime somnolence as an early sign of cognitive decline in a community-based study of older people. Int. J. Geriatr. Psychiatry 2016, 31, 247–255. [Google Scholar] [CrossRef]
- Kyriacou, C.P.; Hastings, M.H. Circadian clocks: Genes, sleep, and cognition. Trends Cogn. Sci. 2010, 14, 259–267. [Google Scholar] [CrossRef]
- Sakai, T.; Tamura, T.; Kitamoto, T.; Kidokoro, Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 16058–16063. [Google Scholar] [CrossRef] [Green Version]
- Guarnieri, B.; Sorbi, S. Sleep and Cognitive Decline: A Strong Bidirectional Relationship. It Is Time for Specific Recommendations on Routine Assessment and the Management of Sleep Disorders in Patients with Mild Cognitive Impairment and Dementia. Eur. Neurol. 2015, 74, 43–48. [Google Scholar] [CrossRef]
- Raber, J. Androgens, apoE, and Alzheimer’s disease. Sci. Aging Knowl. Environ. 2004, 2004, re2. [Google Scholar] [CrossRef]
- Tsapanou, A.; Scarmeas, N.; Gu, Y.; Manly, J.; Schupf, N.; Stern, Y.; Barral, S. Examining the association between Apolipoprotein E (APOE) and self-reported sleep disturbances in non-demented older adults. Neurosci. Lett. 2015, 606, 72–76. [Google Scholar] [CrossRef] [Green Version]
- Tsapanou, A.; Scarmeas, N.; Gu, Y.; Manly, J.; Schupf, N.; Stern, Y.; Barral, S. Data from a cross-sectional study on Apolipoprotein E (APOE-epsilon4) and snoring/sleep apnea in non-demented older adults. Data Brief 2015, 5, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Dashti, H.S.; Jones, S.E.; Wood, A.R.; Lane, J.M.; van Hees, V.T.; Wang, H.; Rhodes, J.A.; Song, Y.; Patel, K.; Anderson, S.G.; et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat. Commun. 2019, 10, 1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, S.R.; Ritchie, S.J.; Allerhand, M.; Hagenaars, S.P.; Radakovic, R.; Breen, D.P.; Davies, G.; Riha, R.L.; Harris, S.E.; Starr, J.M.; et al. Sleep and cognitive aging in the eighth decade of life. Sleep 2019, 42, zsz019. [Google Scholar] [CrossRef] [PubMed]
- Tsapanou, A.; Gao, Y.; Stern, Y.; Barral, S. Polygenic score for sleep duration. Association with cognition. Sleep Med. 2020, 74, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Dardiotis, E.; Kosmidis, M.H.; Yannakoulia, M.; Hadjigeorgiou, G.M.; Scarmeas, N. The Hellenic Longitudinal Investigation of Aging and Diet (HELIAD): Rationale, study design, and cohort description. Neuroepidemiology 2014, 43, 9–14. [Google Scholar] [CrossRef]
- Blessed, G.; Tomlinson, B.E.; Roth, M. The association between quantitative measures of dementia and of senile change in the cerebral grey matter of elderly subjects. Br. J. Psychiatry 1968, 114, 797–811. [Google Scholar] [CrossRef]
- Lawton, M.P.; Brody, E.M. Assessment of older people: Self-maintaining and instrumental activities of daily living. Gerontologist 1969, 9, 179–186. [Google Scholar] [CrossRef]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Lezak, M.D.; Howieson, D.B.; Loring, D.W.; Fischer, J.S. Neuropsychological Assessment; Oxford University Press: New York, NY, USA, 2004. [Google Scholar]
- Vlahou, C.H.; Kosmidis, M.H.; Dardagani, A.; Tsotsi, S.; Giannakou, M.; Giazkoulidou, A.; Zervoudakis, E.; Pontikakis, N. Development of the Greek Verbal Learning Test: Reliability, Construct Validity, and Normative Standards. Arch. Clin. Neuropsychol. 2012, 28, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Kosmidis, M.H.; Vlahou, C.H.; Panagiotaki, P.; Kiosseoglou, G. The verbal fluency task in the Greek population: Normative data, and clustering and switching strategies. J. Int. Neuropsychol. Soc. 2004, 10, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Tsapkini, K.; Vlahou, C.H.; Potagas, C. Adaptation and Validation of Standardized Aphasia Tests in Different Languages: Lessons from the Boston Diagnostic Aphasia Examination–Short Form in Greek. Behav. Neurol. 2010, 22, 423841. [Google Scholar] [CrossRef]
- Benton, A.L.; Sivan, A.B.; Hamsher, K.D.; Varney, N.R.; Spreen, O. Contributions to Neuropsychological Assessment: A Clinical Manual; Oxford University Press: New York, NY, USA, 1994. [Google Scholar]
- Kosmidis, M.H.; Tsotsi, S.; Karambela, O.; Takou, E.; Vlahou, C.H. Cultural Factors Influencing Performance on Visuoperceptual Neuropsychological Tasks. Behav. Neurol. 2010, 23, 956709. [Google Scholar] [CrossRef]
- Bozikas, V.P.; Giazkoulidou, A.; Hatzigeorgiadou, M.; Karavatos, A.; Kosmidis, M.H. Do age and education contribute to performance on the clock drawing test? Normative data for the Greek population. J. Clin. Exp. Neuropsychol. 2008, 30, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Vlahou, C.H.; Kosmidis, M.H. The Greek Trail Making Test: Preliminary norms for clinical and research use. Psychol. J. Hell. Psychol. Soc. 2002, 9, 336–352. (In Greek) [Google Scholar]
- Giaglis, G.; Kyriazidou, S.; Paraskevopoulou, E.; Tascos, N.; Kosmidis, M. Evaluating premorbid level: Preliminary findings regarding the vulnerability of scores on cognitive measures in patients with MS. J. Int. Neuropsychol. Soc. 2010, 15. [Google Scholar]
- Spritzer, K.L.; Hays, R.D. MOS Sleep Scale: A Manual for Use and Scoring, Version 1.0; RAND: Los Angeles, CA, USA, 2003. [Google Scholar]
- Bellenguez, C.; Küçükali, F.; Jansen, I.; Andrade, V.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Grenier-Boley, B.; Campos-Martin, R.; Holmans, P.A.; et al. New insights on the genetic etiology of Alzheimer’s and related dementia. medRxiv 2020. preprint. [Google Scholar] [CrossRef]
- Grove, M.L.; Yu, B.; Cochran, B.J.; Haritunians, T.; Bis, J.C.; Taylor, K.D.; Hansen, M.; Borecki, I.B.; Cupples, L.A.; Fornage, M.; et al. Best practices and joint calling of the HumanExome BeadChip: The CHARGE Consortium. PLoS ONE 2013, 8, e68095. [Google Scholar] [CrossRef]
- Chang, C.C. Data Management and Summary Statistics with PLINK. In Statistical Population Genomics; Dutheil, J.Y., Ed.; Springer: New York, NY, USA, 2020; pp. 49–65. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Abraham, G.; Qiu, Y.; Inouye, M. FlashPCA2: Principal component analysis of Biobank-scale genotype datasets. Bioinformatics 2017, 33, 2776–2778. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Marchini, J.; Howie, B.; Myers, S.; McVean, G.; Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007, 39, 906–913. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, P.-R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; Reshef, Y.A.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altstiel, L.D.; Greenberg, D.A.; Marin, D.; Lantz, M.; Mohs, R. Apolipoprotein E genotype and cognition in the very old. Lancet 1997, 349, 1451. [Google Scholar] [CrossRef]
- Savitz, J.; Solms, M.; Ramesar, R. Apolipoprotein E variants and cognition in healthy individuals: A critical opinion. Brain Res. Rev. 2006, 51, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Mayeux, R.; Ottman, R.; Maestre, G.; Ngai, C.; Tang, M.X.; Ginsberg, H.; Chun, M.; Tycko, B.; Shelanski, M. Synergistic effects of traumatic head injury and apolipoprotein-epsilon 4 in patients with Alzheimer’s disease. Neurology 1995, 45, 555–557. [Google Scholar] [CrossRef]
- Choi, S.W.; O’Reilly, P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 2019, 8, giz082. [Google Scholar] [CrossRef]
- Jansen, P.R.; Watanabe, K.; Stringer, S.; Skene, N.; Bryois, J.; Hammerschlag, A.R.; de Leeuw, C.A.; Benjamins, J.S.; Munoz-Manchado, A.B.; Nagel, M.; et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet. 2019, 51, 394–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarmeas, N.; Stern, Y.; Tang, M.X.; Mayeux, R.; Luchsinger, J.A. Mediterranean diet and risk for Alzheimer’s disease. Ann. Neurol. 2006, 59, 912–921. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, S.A.; Figorilli, M.; Casaceli, G.; Proserpio, P.; Nobili, L. Sleep Related Hypermotor Seizures with a Right Parietal Onset. J. Clin. Sleep Med. 2015, 11, 953–955. [Google Scholar] [CrossRef] [Green Version]
- Lythe, K.E.; Williams, S.C.; Anderson, C.; Libri, V.; Mehta, M.A. Frontal and parietal activity after sleep deprivation is dependent on task difficulty and can be predicted by the fMRI response after normal sleep. Behav. Brain Res. 2012, 233, 62–70. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, N.; Bryant, D.C.; MacLean, J.N.; Gonzalez, C.L.R. Assessing Visuospatial Abilities in Healthy Aging: A Novel Visuomotor Task. Front. Aging Neurosci. 2016, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massaldjieva, R.I. Differentiating normal cognitive aging from cognitive impairment no dementia: A focus on constructive and visuospatial abilities. Gerontology 2018, 9, 167. [Google Scholar]
| Characteristics | |
|---|---|
| Age, years Mean (SD) | 73.1 (5) |
| Sex, women N (%) | 1184 (60.5) |
| Education, years Mean (SD) | 8.4 (4.9) |
| Cognitive domains | |
| Memory, Mean (SD) | −0.1342 (0.94031) |
| Executive, Mean (SD) | −0.1400 (0.78494) |
| Visuo-spatial, Mean (SD) | −0.1963 (0.92621) |
| Language, Mean (SD) | −0.0976 (0.90150) |
| Attention/Speed, Mean (SD) | −0.1840 (1.14814) |
| Duration of follow-up, years Mean (SD) | 3.1 (0.9) |
| Total | 1376 |
| Cognitive Ability | Β * | p |
|---|---|---|
| Memory | −551.69 | 0.173 |
| −759.8 | 0.083 | |
| Executive | −269.87 | 0.417 |
| −161.71 | 0.655 | |
| Visuo-spatial | −1305.22 | 0.018 |
| −1273.59 | 0.031 | |
| Language | 30.947 | 0.936 |
| 68.92 | 0.868 | |
| Attention/Speed | 1253.22 | 0.058 |
| 1184.24 | 0.086 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsapanou, A.; Mourtzi, N.; Charisis, S.; Hatzimanolis, A.; Ntanasi, E.; Kosmidis, M.H.; Yannakoulia, M.; Hadjigeorgiou, G.; Dardiotis, E.; Sakka, P.; et al. Sleep Polygenic Risk Score Is Associated with Cognitive Changes over Time. Genes 2022, 13, 63. https://doi.org/10.3390/genes13010063
Tsapanou A, Mourtzi N, Charisis S, Hatzimanolis A, Ntanasi E, Kosmidis MH, Yannakoulia M, Hadjigeorgiou G, Dardiotis E, Sakka P, et al. Sleep Polygenic Risk Score Is Associated with Cognitive Changes over Time. Genes. 2022; 13(1):63. https://doi.org/10.3390/genes13010063
Chicago/Turabian StyleTsapanou, Angeliki, Niki Mourtzi, Sokratis Charisis, Alex Hatzimanolis, Eva Ntanasi, Mary H. Kosmidis, Mary Yannakoulia, Georgios Hadjigeorgiou, Efthimios Dardiotis, Paraskevi Sakka, and et al. 2022. "Sleep Polygenic Risk Score Is Associated with Cognitive Changes over Time" Genes 13, no. 1: 63. https://doi.org/10.3390/genes13010063
APA StyleTsapanou, A., Mourtzi, N., Charisis, S., Hatzimanolis, A., Ntanasi, E., Kosmidis, M. H., Yannakoulia, M., Hadjigeorgiou, G., Dardiotis, E., Sakka, P., Stern, Y., & Scarmeas, N. (2022). Sleep Polygenic Risk Score Is Associated with Cognitive Changes over Time. Genes, 13(1), 63. https://doi.org/10.3390/genes13010063







