Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Isolation
2.2. Primer Design and Sequencing
2.3. Population Genetic Analyses
2.4. Statistical Analysis
- Y is the trait measured on each animal (BW, BL, BH, ChW, ChD, ChW, HW, and CaC);
- μ is the mean for the trait;
- Genotype is the genotype effect;
- Gender is the gender effect;
- Combination is the combination effect of the gender and genotype;
- ε is the random error and is assumed to be independent, N (0, σ2) distribution.
- Y is the trait measured on each animal (BW, BL, BH, ChW, ChD, ChW, HW, and CaC);
- μ is the mean for the trait;
- Genotype is the genotype effect;
- ε is the random error and is assumed to be independent, N (0, σ2) distribution.
3. Results
3.1. Polymorphism in SSTR1 Gene
3.2. Population Genetics and the Linkage Disequilibrium Analysis
3.3. Association Analysis of Genetic Variants and Haplotypes in SSTR1 with Growth Traits of Hulun Buir Sheep
3.3.1. Association Analysis of SSTR1 with Growth Traits at Birth and 4 Months of Age in Hulun Buir Sheep
3.3.2. Association Analysis of SSTR1 with Growth Traits at 9 Months in Hulun Buir Sheep
3.3.3. Association Analysis of SSTR1 with Growth Traits at 16 Months in Hulun Buir Sheep
3.3.4. Haplotype Association Analysis with Growth Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Zhang, L.; Cao, J.; Wu, M.; Ma, X.; Liu, Z.; Liu, R.; Zhao, F.; Wei, C.; Du, L. Genome-Wide Specific Selection in Three Domestic Sheep Breeds. PLoS ONE 2015, 10, e0128688. [Google Scholar] [CrossRef] [PubMed]
- Al-Mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef]
- La, Y.; Zhang, X.; Li, F.; Zhang, D.; Li, C.; Mo, F.; Wang, W. Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep. Genes 2019, 10, 616. [Google Scholar] [CrossRef] [PubMed]
- Brazeau, P.; Vale, W.; Burgus, R.; Ling, N.; Butcher, M.; Rivier, J.; Guillemin, R. Hypothalamic Polypeptide That Inhibits the Secretion of Immunoreactive Pituitary Growth Hormone. Science 1973, 179, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Quan, F.B.; Desban, L.; Mirat, O.; Kermarquer, M.; Roussel, J.; Koëth, F.; Marnas, H.; Djenoune, L.; Lejeune, F.-X.; Tostivint, H.; et al. Somatostatin 1.1 contributes to the innate exploration of zebrafish larva. Sci. Rep. 2020, 10, 15235. [Google Scholar] [CrossRef] [PubMed]
- Maecke, H.R.; Reubi, J.C. Somatostatin Receptors as Targets for Nuclear Medicine Imaging and Radionuclide Treatment. J. Nucl. Med. 2011, 52, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Finley, J.C.W.; Maderdrut, J.L.; Roger, L.J.; Petrusz, P. The immunocytochemical localization of somatostatin-containing neu-rons in the rat central nervous system. Neuroscience 1981, 6, 2173–2192. [Google Scholar] [CrossRef]
- Anzola, L.K.; Rivera, J.N.; Dierckx, R.A.; Lauri, C.; Valabrega, S.; Galli, F.; Lopez, S.M.; Glaudemans, A.W.J.M.; Signore, A. Value of Somatostatin Receptor Scintigraphy with 99mTc-HYNIC-TOC in Patients with Primary Sjögren Syndrome. J. Clin. Med. 2019, 8, 763. [Google Scholar] [CrossRef]
- Weckbecker, G.; Lewis, I.; Albert, R.; Schmid, H.A.; Hoyer, D.; Bruns, C. Opportunities in somatostatin research: Biological, chemical and therapeutic aspects. Nat. Rev. Drug Discov. 2003, 2, 999–1017. [Google Scholar] [CrossRef]
- Luque, R.M.; Gahete, M.D.; Hochgeschwender, U.; Kineman, R.D. Evidence that endogenous SST inhibits ACTH and ghrelin expression by independent pathways. Am. J. Physiol. Metab. 2006, 291, E395–E403. [Google Scholar] [CrossRef] [PubMed]
- Colturi, T.J.; Unger, R.H.; Feldman, M. Role of circulating somatostatin in regulation of gastric acid secretion, gastrin release, and islet cell function. Studies in healthy subjects and duodenal ulcer patients. J. Clin. Investig. 1984, 74, 417–423. [Google Scholar] [CrossRef]
- Lloyd, K.C.; Amirmoazzami, S.; Friedik, F.; Chew, P.; Walsh, J.H. Somatostatin inhibits gastrin release and acid secretion by activating sst2 in dogs. Am. J. Physiol. Content 1997, 272, G1481–G1488. [Google Scholar] [CrossRef] [PubMed]
- Tulassay, Z. Somatostatin and the Gastrointestinal Tract. Scand. J. Gastroenterol. 1998, 33, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Strowski, M.Z.; Parmar, R.M.; Blake, A.D.; Schaeffer, J.M. Somatostatin Inhibits Insulin and Glucagon Secretion via Two Receptor Subtypes: An in Vitro Study of Pancreatic Islets from Somatostatin Receptor 2 Knockout Mice. Endocrinology 2000, 141, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Debus, N.; Dutour, A.; Vuaroqueaux, V.; Oliver, C.; Ouafik, L. The ovine somatostatin receptor subtype 1 (osst1): Partial cloning and tissue distribution. Domest. Anim. Endocrinol. 2001, 21, 73–84. [Google Scholar] [CrossRef]
- Kailey, B.; van de Bunt, M.; Cheley, S.; Johnson, P.R.; MacDonald, P.E.; Gloyn, A.L.; Rorsman, P.; Braun, M. SSTR2 is the functionally dominant somatostatin receptor in human pancreatic β- and α-cells. American journal of physiology. Endocrinol. Metab. 2012, 303, E1107–E1116. [Google Scholar] [CrossRef]
- Kreienkamp, H.-J.; Akgün, E.; Baumeister, H.; Meyerhof, W.; Richter, D. Somatostatin receptor subtype 1 modulates basal inhibition of growth hormone release in somatotrophs. FEBS Lett. 1999, 462, 464–466. [Google Scholar] [CrossRef]
- Nespeca, M.; Giorgetti, C.; Nobile, S.; Ferrini, I.; Simonato, M.; Verlato, G.; Cogo, P.; Carnielli, V.P. Does Whole-Body Hypothermia in Neonates with Hypoxic–Ischemic Encephalopathy Affect Surfactant Disaturated-Phosphatidylcholine Kinetics? PLoS ONE 2016, 11, e0153328. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Norman, M.; Yang, J.; Magnusson, J.; Kreienkamp, H.-J.; Richter, D.; DeMayo, F.; Brunicardi, F. Alterations in glucose homeostasis in SSTR1 gene-ablated mice. Mol. Cell. Endocrinol. 2006, 247, 82–90. [Google Scholar] [CrossRef]
- Jin, Q.J.; Sun, J.J.; Fang, X.T.; Zhang, C.L.; Yang, L.; Chen, D.X.; Shi, X.Y.; Du, Y.; Lan, X.Y.; Chen, H. Molecular characterization and polymorphisms of the caprine Somatostatin (SST) and SST Receptor 1 (SSTR1) genes that are linked with growth traits. Mol. Biol. Rep. 2010, 38, 3129–3135. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhou, H.; Li, S.; Fang, Q.; Luo, Y.; Hickford, J.G.H. Growth and carcass trait association with variation in the somatostatin receptor 1 (SSTR1) gene in New Zealand Romney sheep. N. Z. J. Agric. Res. 2017, 61, 477–486. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Roychoudhury, A.K. Sampling variances of heterozygosity and genetic distance. Genetics 1974, 76, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2004, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Koller, D.; Rodríguez, M.S.; Zubiaur, P.; Ochoa, D.; Almenara, S.; Román, M.; Romero-Palacián, D.; De Miguel-Cáceres, A.; Martín, S.; Gómez, M.N.; et al. The effects of aripiprazole and olanzapine on pupillary light reflex and its relationship with pharmacogenetics in a randomized multiple-dose trial. Br. J. Clin. Pharmacol. 2020, 86, 2051–2062. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Zhou, Y.; Wang, J.; Yu, X.; Tong, J. Identifying Candidate Genes Involved in the Regulation of Early Growth Using Full-Length Transcriptome and RNA-Seq Analyses of Frontal and Parietal Bones and Vertebral Bones in Bighead Carp (Hypophthalmichthys nobilis). Front. Genet. 2021, 11, 603454. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, K. Genetic diversity of Mycobacterium avium subspecies paratuberculosis and the influence of strain type on infection and pathogenesis: A review. Vet. Res. 2015, 46, 64. [Google Scholar] [CrossRef] [PubMed]
- Single Nucleotide Polymorphisms. In Advanced Structural Safety Studies; Springer: Berlin/Heidelberg, Germany, 2009. [CrossRef]
- Li, H.; Yang, H.; Lv, N.; Ma, C.; Li, J.; Shang, Q. Whole exome sequencing and methylation specific multiplex liga-tion dependent probe amplification applied to identify Angelman syndrome due to paternal uniparental disomy in two unre-lated patients. Mol. Med. Rep. 2019, 20, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, R.; Mo, X.; Jiang, R.; Kong, H.; Hua, W.; Wu, X. Polymorphism of ZBTB17 gene is associated with idiopathic dilated cardiomyopathy: A case control study in a Han Chinese population. Eur. J. Med Res. 2013, 18, 10. [Google Scholar] [CrossRef]
- Koch, E.M. The Effects of Demography and Genetics on the Neutral Distribution of Quantitative Traits. Genetics 2019, 211, 1371–1394. [Google Scholar] [CrossRef] [PubMed]
- Routtu, J.; Hall, M.D.; Albere, B.; Beisel, C.; Bergeron, R.D.; Chaturvedi, A.; Choi, J.-H.; Colbourne, J.; De Meester, L.; Stephens, M.T.; et al. An SNP-based second-generation genetic map of Daphnia magna and its application to QTL analysis of phenotypic traits. BMC Genom. 2014, 15, 1033. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Lehmann, B.D.; Sheng, Q.; Shaver, T.M.; Stricker, T.P.; Pietenpol, J.A.; Shyr, Y. Detection of internal exon deletion with exon Del. BMC Bioinform. 2014, 15, 332. [Google Scholar] [CrossRef] [PubMed]
- Radojevic, V.; Bodmer, D. Expression and localization of somatostatin receptor types 3, 4 and 5 in the wild-type, sstr1 and sstr1/sstr2 knockout mouse cochlea. Cell Tissue Res. 2014, 358, 717–727. [Google Scholar] [CrossRef]
- Liu, X.; Lu, R.; Xia, Y.; Sun, J. Global analysis of the eukaryotic pathways and networks regulated by Salmonella typhimurium in mouse intestinal infection in vivo. BMC Genom. 2010, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; An, T.; Li, M.; Yi, Z.; Li, C.; Sun, X.; Guan, X.; Li, L.; Wang, Y.; Zhang, Y.; et al. The association between early-onset cardiac events caused by neoadjuvant or adjuvant chemotherapy in triple-negative breast cancer patients and some novel autophagy-related polymorphisms in their genomic DNA: A real-world study. Cancer Commun. 2018, 38, 71. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.E.; Mis, S.D.; Jia, K.E.; Wilke, C.O. Reduced mRNA Secondary-Structure Stability Near the Start Codon Indicates Functional Genes in Prokaryotes. Genome Biol. Evol. 2012, 4, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Wei, L. Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC Cancer 2019, 19, 359. [Google Scholar] [CrossRef] [PubMed]
- Taboada, G.F.; Luque, R.M.; Bastos, W.; Guimaraes, R.; Marcondes, J.B.; Chimlii, L.M.C.; Fontes, R.; Mata, P.J.P.; Filho, P.N.; Carvalho, D.P.; et al. Quantitative analysis of soma-tostatin receptor subtype (sstr 1-5) gene expression levels in somatotropinomas and non-function pituitary adenomas. Eur. J. Endocrinol. 2007, 156, 65. [Google Scholar] [CrossRef]
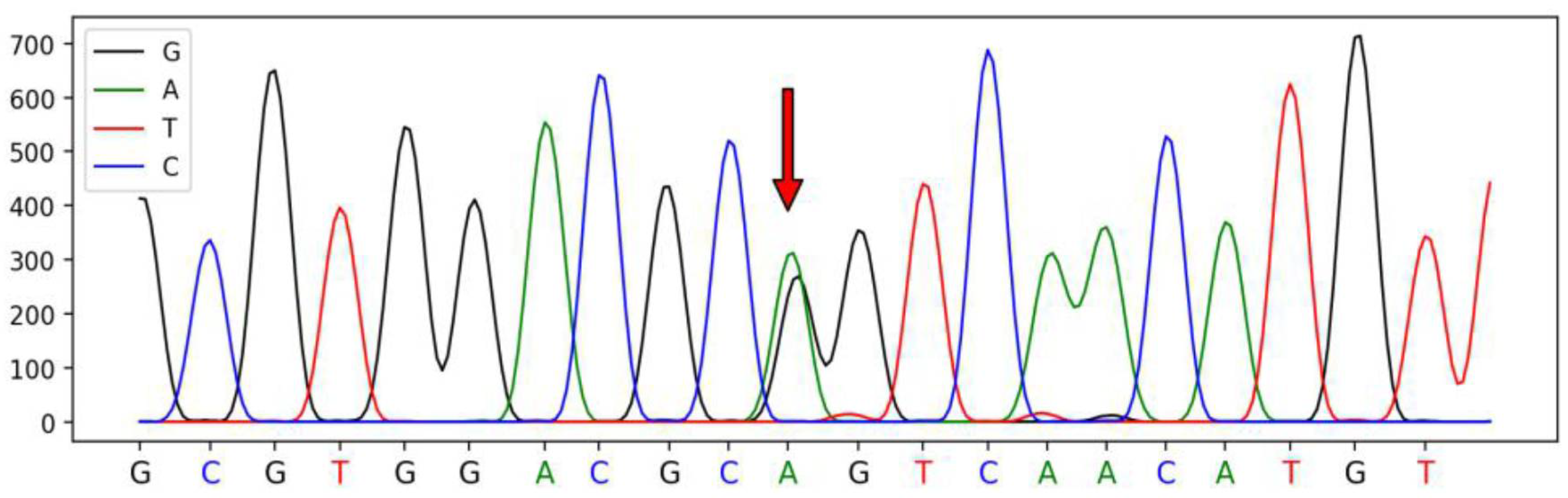
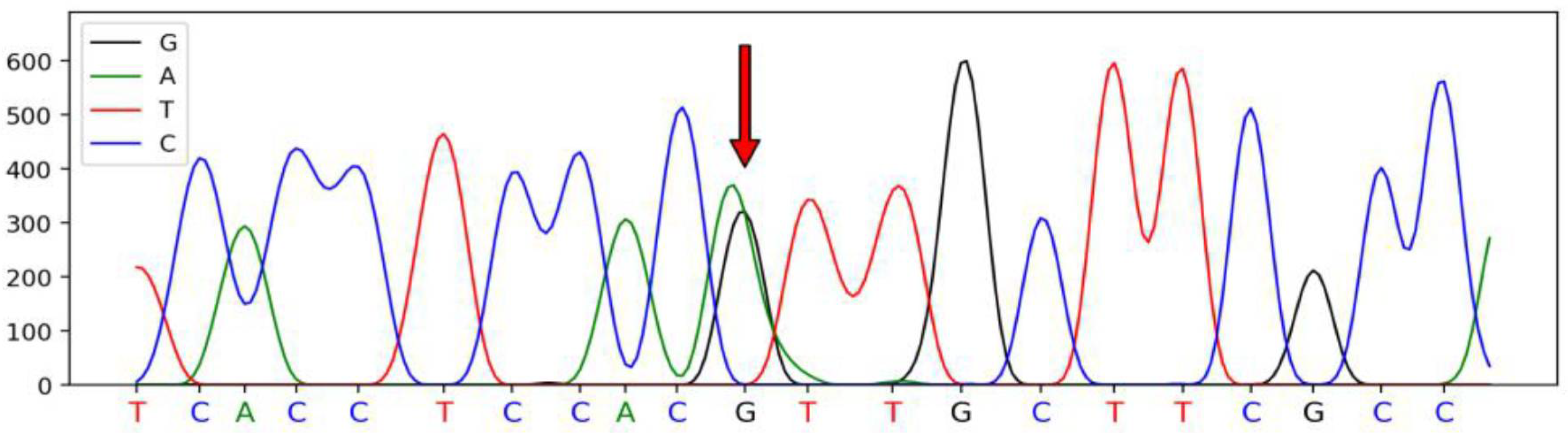
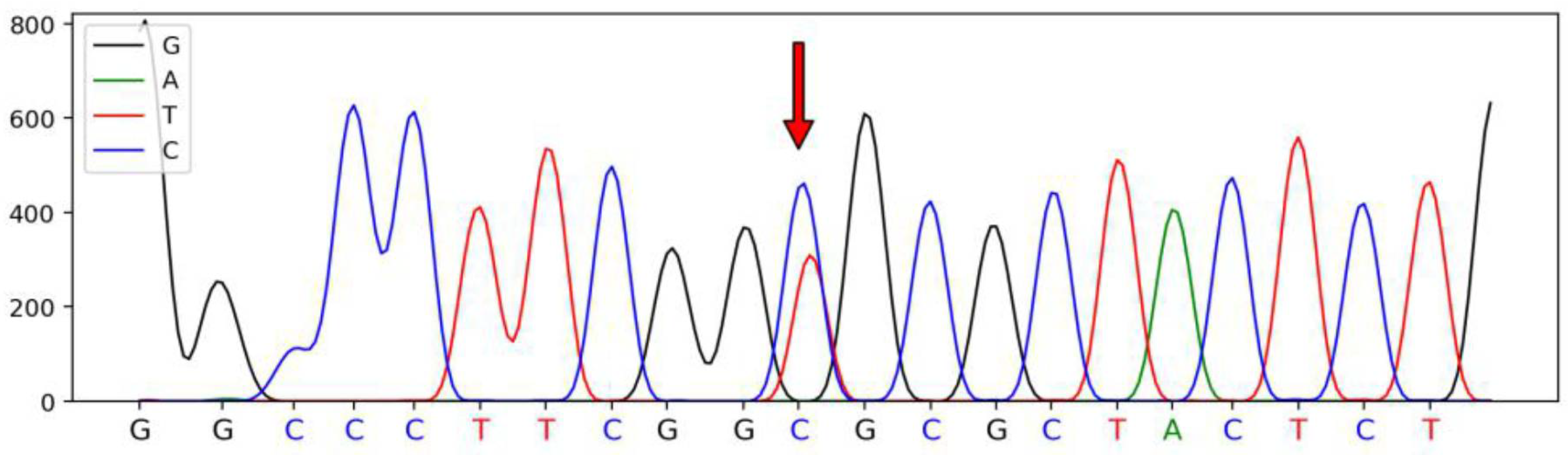


| Primer Names | Primer Sequences (5′–3′) | Size (bp) | Tm (°C) |
|---|---|---|---|
| E1-2 | F:ACATCCTCAACCTGGCCATC | 589 | 60 |
| R:AGCTGCACCACATAGAAGGG | |||
| E1-3 | F:CATGGGCTTCCTGCTGC | 558 | 60 |
| SNP | Allele Frequency | Ho | He | PIC | Ne | HWE | ||
|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | ||||||
| SNP1(A/G) | 0.8233 | 0.1767 | 0 | 0.1612 | 0.1767 | 0.1482 | 1.2146 | 0.2988 |
| SNP2(A/G) | 0.9485 | 0.0515 | 0 | 0.0472 | 0.0503 | 0.0490 | 1.0530 | 0.1198 |
| SNP3(C/T) | 0.8534 | 0.1466 | 0 | 0.1466 | 0.1359 | 0.1266 | 1.1573 | 0.2621 |
| SNP4(G/C) | 0.5193 | 0.4034 | 0.0773 | 0.4034 | 0.4023 | 0.3207 | 1.6731 | 0.5920 |
| SNP | Genotype Frequency | BRW/kg | BW/kg | BL/cm | BH/cm | ChC/cm | ChD/cm | ChW/cm | HW/cm | CaC/cm |
|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 | AA (n = 191) | 4.20 ± 0.05 | 23.47 ± 0.51 | 57.26 ± 0.50 | 55.98 ± 0.39 | 68.44 ± 0.54 | 28.07 ± 0.22 | 15.83 ± 0.17 | 12.48 ± 0.11 | 7.47 ± 0.04 |
| AG (n = 41) | 4.24 ± 0.11 | 22.99 ± 1.11 | 56.95 ± 1.10 | 56.17 ± 0.86 | 68.35 ± 1.18 | 28.24 ± 0.48 | 15.54 ± 0.36 | 12.31 ± 0.23 | 7.49 ± 0.09 | |
| GG (n = 0) | / | / | / | / | / | / | / | / | / | |
| SNP2 | AA (n = 221) | 4.21 ± 0.05 | 23.65 ± 0.47 a | 57.39 ± 0.47 | 56.21 ± 0.36 a | 68.64 ± 0.50 | 28.24 ± 0.20 A | 15.74 ± 0.15 | 12.42 ± 0.10 | 7.49 ± 0.04 |
| AG (n = 12) | 4.18 ± 0.20 | 19.28 ± 2.03 b | 53.87 ± 2.01 | 52.81 ± 1.56 b | 65.31 ± 2.16 | 25.88 ± 0.87 B | 16.68 ± 0.67 | 13.13 ± 0.43 | 7.27 ± 0.17 | |
| GG (n = 0) | / | / | / | / | / | / | / | / | / | |
| SNP3 | CC (n = 198) | 4.23 ± 0.05 | 23.68 ± 0.50 | 57.41 ± 0.49 | 56.20 ± 0.38 | 68.79 ± 0.53 | 28.18 ± 0.21 | 15.91 ± 0.16 | 12.52 ± 0.10 | 7.49 ± 0.04 |
| CT (n = 34) | 4.10 ± 0.12 | 21.96 ± 1.20 | 55.86 ± 1.19 | 55.16 ± 0.92 | 66.68 ± 1.27 | 27.70 ± 0.52 | 15.17 ± 0.39 | 12.07 ± 0.25 | 7.41 ± 0.10 | |
| TT (n = 0) | / | / | / | / | / | / | / | / | / | |
| SNP4 | CC (n = 121) | 4.16 ± 0.06 | 22.93 ± 0.64 | 56.56 ± 0.63 | 55.60 ± 0.49 | 67.86 ± 0.67 | 27.94 ± 0.27 | 15.66 ± 0.21 | 12.45 ± 0.13 | 7.44 ± 0.05 |
| CG (n = 94) | 4.27 ± 0.07 | 23.91 ± 0.73 | 57.81 ± 0.72 | 56.66 ± 0.56 | 69.00 ± 0.77 | 28.20 ± 0.31 | 15.99 ± 0.24 | 12.46 ± 0.15 | 7.53 ± 0.06 | |
| GG (n = 18) | 4.18 ± 0.16 | 24.06 ± 1.66 | 58.39 ± 1.63 | 55.73 ± 1.27 | 69.72 ± 1.75 | 28.89 ± 0.71 | 15.69 ± 0.54 | 12.46 ± 0.35 | 7.41 ± 0.14 |
| SNP | Genotype Frequency | BW/kg | BL/cm | BH/cm | ChC/cm | ChD/cm | ChW/cm | HW/cm | CaC/cm |
|---|---|---|---|---|---|---|---|---|---|
| SNP1 | AA (n = 191) | 32.33 ± 0.55 | 66.51 ± 0.37 | 63.59 ± 0.31 | 83.11 ± 0.57 | 32.64 ± 0.22 | 21.55 ± 0.19 | 14.69 ± 0.12 | 7.56 ± 0.04 |
| AG (n = 41) | 31.77 ± 1.24 | 67.32 ± 0.84 | 64.32 ± 0.70 | 84.07 ± 1.29 | 32.55 ± 0.49 | 21.99 ± 0.42 | 14.42 ± 0.26 | 7.59 ± 0.10 | |
| GG (n = 0) | / | / | / | / | / | / | / | / | |
| SNP2 | AA (n = 221) | 32.52 ± 0.51 a | 66.83 ± 0.35 a | 63.84 ± 0.29 | 83.52 ± 0.53 a | 32.81 ± 0.20 A | 21.58 ± 0.18 | 14.70 ± 0.11 a | 7.58 ± 0.04 |
| AG (n = 12) | 27.84 ± 2.18 b | 63.84 ± 1.47 b | 61.68 ± 1.23 | 78.90 ± 2.26 b | 29.70 ± 0.85 B | 22.46 ± 0.75 | 13.74 ± 0.46 b | 7.31 ± 0.17 | |
| GG (n = 0) | / | / | / | / | / | / | / | / | |
| SNP3 | CC (n = 198) | 32.50 ± 0.54 | 66.87 ± 0.37 | 63.96 ± 0.30 a | 83.44 ± 0.57 | 32.74 ± 0.22 | 21.72 ± 0.19 | 14.67 ± 0.11 | 7.59 ± 0.04 |
| CT (n = 34) | 31.07 ± 1.29 | 65.64 ± 0.87 | 62.41 ± 0.72 b | 82.39 ± 1.36 | 32.07 ± 0.52 | 21.17 ± 0.44 | 14.50 ± 0.27 | 7.48 ± 0.10 | |
| TT (n = 0) | / | / | / | / | / | / | / | / | |
| SNP4 | CC (n = 121) | 31.87 ± 0.69 | 66.43 ± 0.47 | 63.07 ± 0.38 | 82.57 ± 0.72 | 32.34 ± 0.27 b | 21.34 ± 0.23 b | 14.56 ± 0.15 | 7.57 ± 0.05 b |
| CG (n = 94) | 32.59 ± 0.80 | 66.84 ± 0.54 | 64.33 ± 0.45 | 83.92 ± 0.83 | 32.76 ± 0.32 b | 21.77 ± 0.27 b | 14.73 ± 0.17 | 7.49 ± 0.06 b | |
| GG (n = 18) | 33.03 ± 1.78 | 67.36 ± 1.20 | 65.03 ± 0.99 | 84.65 ± 1.84 | 34.03 ± 0.70 a | 22.88 ± 0.60 a | 14.78 ± 0.37 | 7.94 ± 0.14 a |
| SNP | Genotype Frequency | BW/kg | BL/cm | BH/cm | ChC/cm | ChD/cm | ChW/cm | HW/cm | CaC/cm |
|---|---|---|---|---|---|---|---|---|---|
| SNP1 | AA (n = 191) | 38.01 ± 0.48 | 72.61 ± 0.53 | 67.35 ± 0.37 | 81.78 ± 0.40 | 33.60 ± 0.34 | 19.44 ± 0.25 | 18.01 ± 0.16 | 8.18 ± 0.03 |
| AG (n = 41) | 39.19 ± 1.03 | 73.87 ± 1.14 | 67.22 ± 0.79 | 82.36 ± 0.85 | 34.10 ± 0.42 | 19.25 ± 0.50 | 18.42 ± 0.33 | 8.13 ± 0.06 | |
| GG (n = 0) | / | / | / | / | / | / | / | / | |
| SNP2 | AA (n = 221) | 38.48 ± 0.45 | 72.83 ± 0.50 | 67.42 ± 0.34 | 82.06 ± 0.37 | 32.77 ± 0.18 | 19.41 ± 0.22 | 18.13 ± 0.15 | 8.19 ± 0.03 |
| AG (n = 12) | 35.00 ± 1.81 | 72.85 ± 2.00 | 66.11 ± 1.38 | 79.57 ± 1.49 | 32.48 ± 0.74 | 19.07 ± 0.87 | 17.43 ± 0.59 | 7.99 ± 0.11 | |
| GG (n = 0) | / | / | / | / | / | / | / | / | |
| SNP3 | CC (n = 198) | 38.63 ± 0.47 a | 73.32 ± 0.51 A | 67.81 ± 0.35 A | 82.23 ± 0.38 a | 33.80 ± 0.19 | 19.44 ± 0.22 | 18.14 ± 0.15 | 8.18 ± 0.03 |
| CT (n = 34) | 35.94 ± 1.21 b | 69.77 ± 1.32 B | 64.39 ± 0.90 B | 79.85 ± 1.00 b | 33.00 ± 0.50 | 19.14 ± 0.58 | 17.79 ± 0.39 | 8.14 ± 0.07 | |
| TT (n = 0) | / | / | / | / | / | / | / | / | |
| SNP4 | CC (n = 121) | 38.36 ± 0.60 | 72.35 ± 0.66 | 67.06 ± 0.46 | 81.73 ± 0.50 | 33.71 ± 0.24 | 19.34 ± 0.29 | 18.08 ± 0.19 | 8.18 ± 0.04 |
| CG (n = 94) | 38.13 ± 0.70 | 73.66 ± 0.76 | 67.69 ± 0.53 | 82.13 ± 0.57 | 33.83 ± 0.28 | 19.57 ± 0.33 | 18.22 ± 0.23 | 8.17 ± 0.04 | |
| GG (n = 18) | 38.38 ± 1.62 | 71.74 ± 1.76 | 67.55 ± 1.22 | 82.07 ± 1.33 | 32.87 ± 0.65 | 18.77 ± 0.77 | 17.41 ± 0.52 | 8.14 ± 0.10 |
| Genotypes | BRW/kg | BW/kg | BL/cm | BH/cm | ChC/cm | ChD/cm | ChW/cm | HW/cm | CaC/cm |
|---|---|---|---|---|---|---|---|---|---|
| ACC (n = 176) | 4.23 ± 0.05 | 23.13 ± 0.53 | 57.01 ± 0.57 | 56.04 ± 0.42 | 68.12 ± 0.56 | 27.97 ± 0.23 | 15.72 ± 0.16 | 12.49 ± 0.11 | 7.48 ± 0.04 |
| ACG (n = 88) | 4.25 ± 0.07 | 23.74 ± 0.75 | 57.93 ± 0.80 | 56.33 ± 0.60 | 68.88 ± 0.79 | 28.21 ± 0.33 | 15.86 ± 0.23 | 12.50 ± 0.16 | 7.50 ± 0.06 |
| ATC (n = 25) | 4.13 ± 0.14 | 20.48 ± 1.40 | 55.02 ± 1.50 | 54.30 ± 1.12 | 65.22 ± 1.49 | 27.74 ± 0.62 | 14.54 ± 0.77 | 12.06 ± 0.29 | 7.30 ± 0.11 |
| ATG (n = 8) | 4.30 ± 0.25 | 23.16 ± 2.47 | 58.81 ± 2.65 | 55.69 ± 1.98 | 67.19 ± 3.68 | 28.38 ± 1.10 | 14.94 ± 0.77 | 12.00 ± 0.51 | 7.50 ± 0.20 |
| GCC (n = 36) | 4.25 ± 0.12 | 23.17 ± 1.17 | 57.60 ± 1.25 | 56.39 ± 0.93 | 68.33 ± 1.24 | 28.46 ± 0.52 | 15.44 ± 0.36 | 12.26 ± 0.24 | 7.45 ± 0.10 |
| GCG (n = 11) | 4.22 ± 0.21 | 26.10 ± 2.11 | 60.96 ± 2.26 | 58.73 ± 1.69 | 72.00 ± 2.24 | 29.23 ± 0.94 | 16.36 ± 0.65 | 12.55 ± 0.44 | 7.62 ± 0.17 |
| GTC (n = 3) | 4.06 ± 0.40 | 17.40 ± 4.04 | 51.83 ± 4.33 | 52.33 ± 3.24 | 64.83 ± 4.29 | 27.17 ± 1.79 | 15.17 ± 1.25 | 12.50 ± 0.84 | 7.00 ± 0.33 |
| p | 0.987 | 0.223 | 0.258 | 0.357 | 0.211 | 0.805 | 0.123 | 0.753 | 0.512 |
| Genotypes | BW/kg | BL/cm | BH/cm | ChC/cm | ChD/cm | ChW/cm | HW/cm | CaC/cm |
|---|---|---|---|---|---|---|---|---|
| ACC (n = 176) | 31.98 ± 0.57 | 66.58 ± 0.39 | 63.66 ± 0.33 | 82.90 ± 0.63 | 32.48 ± 0.23 | 21.45 ± 0.20 | 14.56 ± 0.12 | 7.53 ± 0.05 |
| ACG (n = 88) | 32.60 ± 0.81 | 67.10 ± 0.55 | 64.42 ± 0.47 | 84.07 ± 0.89 | 32.98 ± 0.32 | 22.10 ± 0.28 | 14.66 ± 0.16 | 7.56 ± 0.06 |
| ATC (n = 25) | 29.92 ± 1.51 | 64.54 ± 1.02 | 62.20 ± 0.88 | 80.88 ± 1.67 | 31.65 ± 0.59 | 20.96 ± 0.53 | 14.23 ± 0.31 | 7.48 ± 0.12 |
| ATG (n = 8) | 32.16 ± 2.67 | 66.63 ± 1.81 | 64.00 ± 1.56 | 83.94 ± 2.95 | 32.50 ± 1.07 | 21.13 ± 0.94 | 14.81 ± 0.55 | 7.25 ± 0.21 |
| GCC (n = 36) | 32.12 ± 1.26 | 67.57 ± 0.85 | 63.85 ± 0.74 | 84.40 ± 1.39 | 32.75 ± 0.51 | 22.25 ± 0.44 | 14.33 ± 0.26 | 7.67 ± 0.10 |
| GCG (n = 11) | 34.70 ± 2.28 | 69.86 ± 1.54 | 65.55 ± 1.33 | 87.96 ± 2.52 | 34.41 ± 0.91 | 22.82 ± 0.80 | 14.73 ± 0.47 | 7.64 ± 0.28 |
| GTC (n = 3) | 26.67 ± 4.36 | 64.00 ± 2.95 | 63.00 ± 2.55 | 76.33 ± 4.82 | 31.00 ± 1.75 | 22.00 ± 1.53 | 13.00 ± 0.90 | 7.50 ± 0.35 |
| p | 0.502 | 0.087 | 0.325 | 0.154 | 0.225 | 0.152 | 0.416 | 0.585 |
| Genotypes | BW/kg | BL/cm | BH/cm | ChD/cm | HW/cm | CaC/cm |
|---|---|---|---|---|---|---|
| ACC (n = 176) | 38.21 ± 0.46 | 72.44 ± 0.60 | 67.34 ± 0.34 | 33.97 ± 0.34 | 18.10 ± 0.16 | 8.23 ± 0.06 |
| ACG (n = 88) | 38.13 ± 0.64 | 73.24 ± 0.84 | 66.64 ± 0.49 | 33.53 ± 0.47 | 18.06 ± 0.22 | 8.16 ± 0.09 |
| ATC (n = 25) | 35.73 ± 1.19 | 69.19 ± 1.57 | 64.12 ± 0.90 | 32.92 ± 0.88 | 17.67 ± 0.41 | 8.14 ± 0.16 |
| ATG (n = 8) | 36.26 ± 2.15 | 70.38 ± 2.82 | 65.63 ± 1.63 | 33.38 ± 1.59 | 17.69 ± 0.73 | 8.13 ± 0.29 |
| GCC (n = 36) | 39.09 ± 1.01 | 72.01 ± 1.33 | 67.11 ± 0.77 | 35.25 ± 0.75 | 18.04 ± 0.35 | 8.38 ± 0.14 |
| GCG (n = 11) | 39.69 ± 1.84 | 75.09 ± 2.41 | 68.09 ± 1.39 | 34.18 ± 1.35 | 18.27 ± 0.63 | 8.18 ± 0.25 |
| GTC (n = 3) | 35.73 ± 3.51 | 74.00 ± 4.61 | 64.67 ± 2.65 | 33.33 ± 2.59 | 18.67 ± 1.20 | 8.00 ± 0.48 |
| p | 0.291 | 0.312 | 0.395 | 0.498 | 0.970 | 0.869 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Ding, N.; Zhang, Z.; Tian, D.; Han, B.; Liu, S.; Liu, D.; Tian, F.; Zhao, K. Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep. Genes 2022, 13, 77. https://doi.org/10.3390/genes13010077
Li X, Ding N, Zhang Z, Tian D, Han B, Liu S, Liu D, Tian F, Zhao K. Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep. Genes. 2022; 13(1):77. https://doi.org/10.3390/genes13010077
Chicago/Turabian StyleLi, Xue, Ning Ding, Zhichao Zhang, Dehong Tian, Buying Han, Sijia Liu, Dehui Liu, Fei Tian, and Kai Zhao. 2022. "Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep" Genes 13, no. 1: 77. https://doi.org/10.3390/genes13010077
APA StyleLi, X., Ding, N., Zhang, Z., Tian, D., Han, B., Liu, S., Liu, D., Tian, F., & Zhao, K. (2022). Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep. Genes, 13(1), 77. https://doi.org/10.3390/genes13010077






