Abstract
The heat shock protein 70 (HSP70) gene family perform a fundamental role in protecting plants against biotic and abiotic stresses. Aquilaria sinensis is a classic stress-induced medicinal plant, producing a valuable dark resin in a wood matrix, known as agarwood, in response to environmental stresses. The HSP70 gene family has been systematic identified in many plants, but there is no comprehensive analysis at the genomic level in A. sinensis. In this study, 15 putative HSP70 genes were identified in A. sinensis through genome-wide bioinformatics analysis. Based on their phylogenetic relationships, the 15 AsHSP70 were grouped into six sub-families that with the conserved motifs and gene structures, and the genes were mapped onto six separate linkage groups. A qRT-PCR analysis showed that the relative expression levels of all the AsHSP70 genes were up-regulated by heat stress. Subcellular localization of all HSP70s was predicted, and three were verified by transiently expressed in Arabidopsis protoplasts. Based on the expression profiles in different tissues and different layers treated with Agar-Wit, we predict AsHSP70 genes are involved in different stages of agarwood formation. The systematic identification and expression analysis of HSP70s gene family imply some of them may play important roles in the formation of agarwood. Our findings not only provide a foundation for further study their biological function in the later research in A. sinensis, but also provides a reference for the analysis of HSPs in other species.
1. Introduction
Plants are exposed to dynamic and complex environmental stimuli during their growth, including not only biotic stresses, such as herbivore and pathogen attacks, but also abiotic stresses, such as extreme temperatures, salinity, chilling and drought, causing cell injures and producing secondary stresses. Over the course of long-term evolution, plants have evolved several mechanisms, morphological and physiological adaptations to protect themselves [1,2]. Heat-shock proteins (HSP) are stressed-related proteins and could be induced by a broad spectrum of environmental stresses, such as heat [3], drought [4], salinity [5] and heavy metals [6,7,8]. Since they were first discovered in Drosophila in the 1960s, HSPs have been identified in almost all organisms [9]. According to their molecular weight, HSP superfamily proteins are classified into the following five families: HSP60, chaperonin family; HSP70, 70-kDa-heat shock protein; HSP90 family; HSP100 family; sHSP, small heat shock protein [2]. Among these families, HSP70s have drawn the greatest attention as they have housekeeping functions in protein folding and protein quality control by proofreading protein structure and repairing misfolded conformers, preventing protein aggregation or misfolded conformers [10]. Structurally, HSP70s are comprised of the following three major domains: a conserved 44-kD N-terminal ATPase domain (NBD), an 18-kD substrate binding domain (SBD) and a 10-kD variable C-terminal domain. HSP70 are considered to be the most highly conserved HSPs evolutionarily [2,11,12,13].
Diverse biological functions of HSP70 gene family have been well characterized in several plant systems. Numerous studies have reported HSP70s play vital roles in stress response. For example, it is demonstrated that high expression of HSP70s led to tolerance enhancement in several plants [14,15]. In Nicotiana tabacum, mutant plants with overexpression of NtHSP70-1 gene showed high tolerance to drought stress [16]. HSP70s also function with drought and salt tolerance in Saccharum spp. Hybrid [17], Dunaliella salina [18] and Sorghum bicolor [19]. Moreover, HSP70 developmental expression appears to be complex during the growth of vegetative and reproductive stage. Transgenic cpHSC70-1 in Arabidopsis exhibited malformed leaves, variegated cotyledons, impaired root growth, and growth retardation, after heat stress treatment of germinating seeds [20]. In Arabidopsis, the cpHSP70s are also essential for maintaining chloroplast development. The cpHsc70-1/cpHsc70-2 double-mutants show a white and stunted phenotype [21]. HSP70s have been well identified in many plant species. So far, 18 copies of HSP70s have been reported in Arabidopsis [22]. Additionally, 16 members in Chenopodium quinoa [23], 18 members in Pennisetum glaucum [24] and 61 putative HSP70 genes in Glycine max L. [25] are also reported.
Aquilaria sinensis is a typical stress-induced medicinal plant responding to stress with the formation of dark resinous wood called agarwood, which is widely used in traditional medicine as a digestive, sedative, and antiemetic, as well as being used as a precious scent and perfume across Asia, the Middle East and Europe [26,27,28]. Agarwoods are generally formed only in naturally or artificially injured trees [29,30], and high-quality agarwood is more valuable than gold on a unit weight basis, in the international market. According to the China Pharmacopoeia, A. sinensis is the only certified source for agarwood [31]. Agarwood formation and accumulation in natural conditions are infrequent and take over decades. Though trees are widespread in South China and Southeast Asia, millions of Aquilaria trees are unable to produce agarwood, which makes it a very scarce material. The successful development of a reliable agarwood induction protocol will necessitate a better understanding of the wound-induction molecular mechanism. Our group has previously verified that agarwood formation is the production of defense response in A. sinensis [32,33,34,35,36]. HSP70s are a highly conserved protein and perform a fundamental role in protecting plants against abiotic stresses, which means they might play potential function in agarwood formation. However, to date, the HSP70 gene family members in A. sinensis have not been reported. Thus, it is necessary to analyze the protein family to obtain a better understanding of the stress damage defensive mechanism of A. sinensis.
In this study, a total of 15 HSP70 genes were identified using a bioinformatic method based on the genome of A. sinensis. Comprehensive analyses were conducted to reveal the gene chromosomal locations, phylogenetic relationships, gene structures and conserved motifs. Moreover, the expression profiles of the AsHSP70 family in response to heat stress were revealed by the qRT-PCR, and the subcellular localization were studied and verified by transient expression in Arabidopsis protoplasts. In addition, the expression patterns of AsHSP70 genes were detected in different tissues and Agar-Wit treatment in A. sinensis.
2. Materials and Methods
2.1. Identification of HSP70s in Plant Species
To obtain potential members of the HSP70 gene family in A. sinensis, the amino acid sequences of the HSP70s of Arabidopsis were downloaded from the TAIR (http://www.arabidopsis.org/, accessed on 28 March 2021), rice HSP70 protein sequences were obtained from the TIGR Database, and the sequences of Marchantia polymorpha, Selaginella moellendorffii, Amborella trichopoda, G. max, Picea abies, Hordeum vulgare were downloaded from NCBI databases. The eight plant species HSP70 protein sequences were used as queries in basic local alignment search tool (BLATP) search of the A. sinensis genome database with the criteria of an e-value < 10−5. The results were scanned with the identity ≥50 and align ratio ≥ 50%. A total of 15 A. sinensis HSP70 gene sequences were obtained after manually filtering out repeated sequences, and sequences without HSP70 domains by HMMSCAN.
2.2. Phylogenetic Analyses
Based on the HSP70 protein sequences, ClustalW was used to simultaneously align, the phylogenetic tree was constructed using MEGA7.0 with the Neighbor-Joining (NJ) method, and a bootstrap analysis was conducted using 1000 replicates at each node.
2.3. Gene Structure Analysis, Conserved Motif Analysis and Chromosomal Location Analysis
To investigate the AsHSP70 genes diversity and structure, exon and intron structures of AsHSP70 genes were illustrated using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn, accessed on 15 June 2021) [37]. The information of AsHSP70 proteins conserved motifs were identified using the MEME program (version 4.11.2, http://alternate.memesuite.org/tools/meme, accessed on 18 July 2021). The chromosomal locations of the AsHSP70 genes were determined using the Populus genome browser (http://www.phytozome.net/poplar, accessed on 16 June 2021). WoLF PSORT software was used to predict the subcellular localization of AsHSP70 proteins based on the chromosomal position information provided in the genomic annotation file [38]. The molecular weight (Da) and isoelectric point (pI) of each gene were obtained using compute pI/Mw tool from ExPASy (http://www.expasy.org/tools/, accessed on 20 July 2021) [39]. The instability index, aliphatic index and Grand average of hydropathicity of the identified proteins were calculated by using ProtParam Tool (http://web.expasy.org/protparam, accessed on 20 July 2021).
2.4. Plant Materials and Stress Treatments
A. sinensis calli were used as material in this study. All the lines were cultured in dark at 25 °C. For the heat stress, A. sinensis calli were removed to a high temperature chamber (42 °C) for 0 h, 6 h, 12 h, 24 h, 36 h, 48 h and 72 h and the calli without any treatment were used as the control. All materials were frozen in liquid nitrogen and stored at −80 °C for further qRT-PCR analysis. The materials for transcription sequence are stems from seven-year-old A. sinensis; the trees are grown in wild field in Hainan and were treated using Whole-tree agarwood-inducing technique (Agar-Wit) [40]. Samples were collected at different times. The healthy wood was used as control.
2.5. Subcellular Localization in Arabidopsis Protoplasts
The CDS sequences of AsHSP70-4, AsHSP70-5, AsHSP70-7 and AsHSP70-14 were PCR-amplified with the gene-specific primers, the amplification products were digested with BamHI and Encore, then cloned into the pBWA(V)HS-GFP plasmid, resulting in recombinant expression vectors of AsHSP70s with the CDS of enhanced green fluorescent protein (eGFP) protein. The fusion plasmids were transformed into Arabidopsis protoplasts as Kim has been reported previously [41]. The pBWA(V)HS-GFP empty vector served as the positive control. Images were acquired at 48 h using a Leica DMLe camera (Leica, Wetzlar, Germany).
2.6. RNA Isolation and Real-Time qRT-PCR
The total RNA was extracted from treated calli using a Total RNA Rapid Extraction kit RN38-EASYspin Plus (Aidlab, Gdańsk, Poland). An amount of 1 μg of total RNA was reverse-transcribed to cDNA using the PrimeScript™ RT Reagent Kit (Takara, Dalian, China) according to the manufacturer’s protocol. The PCR amplifications were performed using SYBR® Premix Ex Taq™ II (Takara, Dalian, China) on Light Cycler® 480II (Roche Diagnostics, Indianapolis, IN, USA). Each reaction contained 5 μL SYBR® Premix Ex Taq II, 3 μL ddH2O, 1 μL cDNA template, and 0.5 μL gene-specific primer in a final volume of 10 μL. The PCR cycling conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Three independent biological replicates were performed, and the relative expression level for each of AsHSP70s was calculated using the 2−ΔΔCT method [42].
3. Results
3.1. Identification of the HSP70 Gene Family in A. sinensis
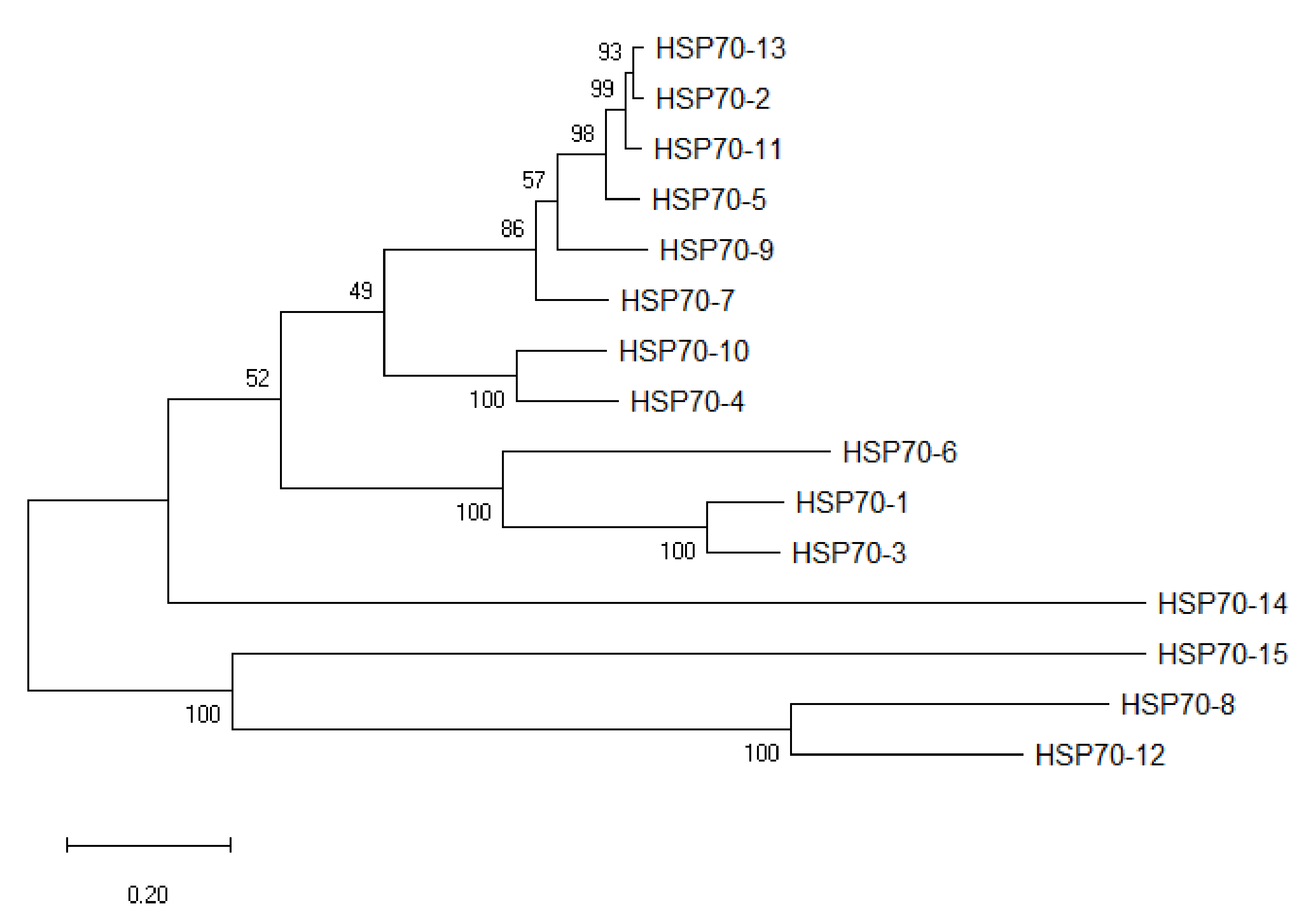
HSP70 sequences of eight plant species were used as queries in BLASTP against protein database in A. sinensis with a maximum E-value of 1 × 10−5. A total of 15 putative HSP70 gene sequences of A. sinensis were finally obtained and named based on the order of their location on the chromosomes. The AsHSP70s encoded proteins varied from 489 to 893 amino acids (aa) in length. Among these proteins, AsHSP70-7 protein sequence was the shortest one with 489 amino acids, and AsHSP70-15 encoded the longest protein with 893 amino acids. The predicted molecular weight of AsHSP70 protein range from 5.3 KD (AsHSP70-7, 489 aa) to 10 KD (AsHSP70-15, 893 aa) and the predicted isoelectric points (PI) values were between 5.12 (AsHSP70-12) and 5.83 (AsHSP70-1). Based on the ExPASy analysis, the predicted protein instability indices showed 3 of the 15 AsHSP70 proteins could be considered as unstable proteins, and the other 12 AsHSP70 proteins were predicted to be stable (cutoff < 40). The detailed information of all AsHSP70 genes is shown in Table 1, and the figure after the localization showed the different possibility of subcellular localization for proteins. To classify these 15 AsHSP70s, the neighbor-joining tree was constructed based on the AsHSP70 protein sequences (Figure 1).

Table 1.
Summary information of predicted physiological and biochemical properties of the AsHSP70 proteins based on the ExPASy analysis.
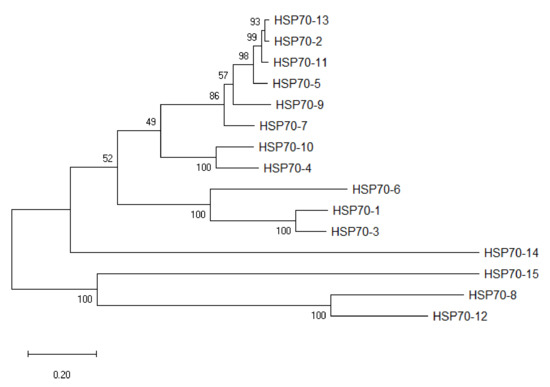
Figure 1.
Phylogenetic tree of AsHSP70s in A. sinensis. Fifteen AsHSP70 members were identified from the genome in A. sinensis. Unrooted phylogenetic tree was constructed based on multiply-aligned sequences of the 15 AsHSP70 proteins with 1000 bootstrap replications.
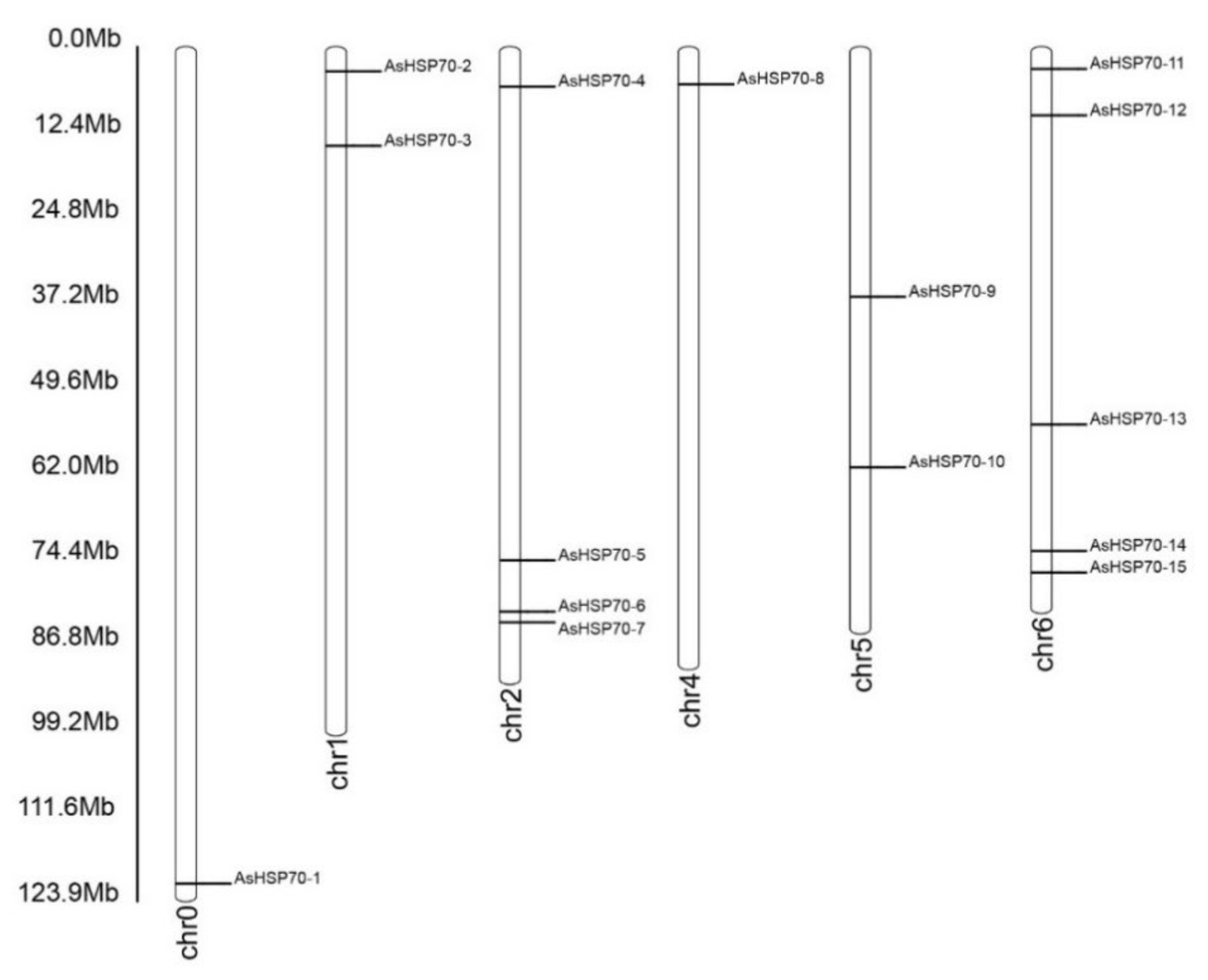
An analysis of the chromosomal location showed that fifteen putative AsHSP70 genes were distributed randomly on six chromosomes and the number on each chromosome was independent to chromosome length (Figure 2). The majority of AsHSP70 genes appeared to congregate at the proximate or the distal ends of the chromosomes. Five predicted AsHSP70 genes were present on chr 6, followed by four genes in chr 2. Only one gene was present in chr 0 and chr 4 with the lowest density. Two genes each were found in chr 6, chr 1 and chr 5.
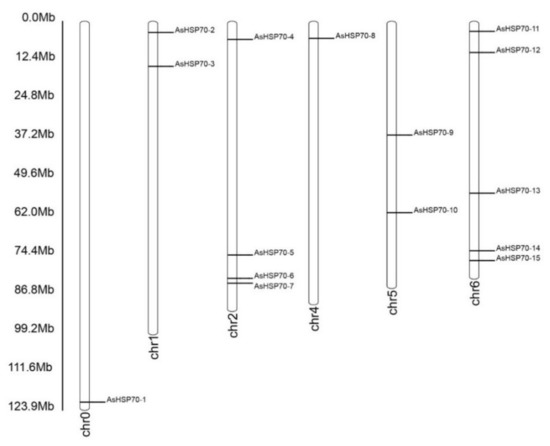
Figure 2.
Chromosomal location of AsHSP70 in A. sinensis. The chromosome number is indicated at the bottom of each chromosome. The bar located on the left side indicates the chromosome sizes in mega bases (Mb).
3.2. Phylogenetic Relationship of the HSP70 Genes in A. sinensis
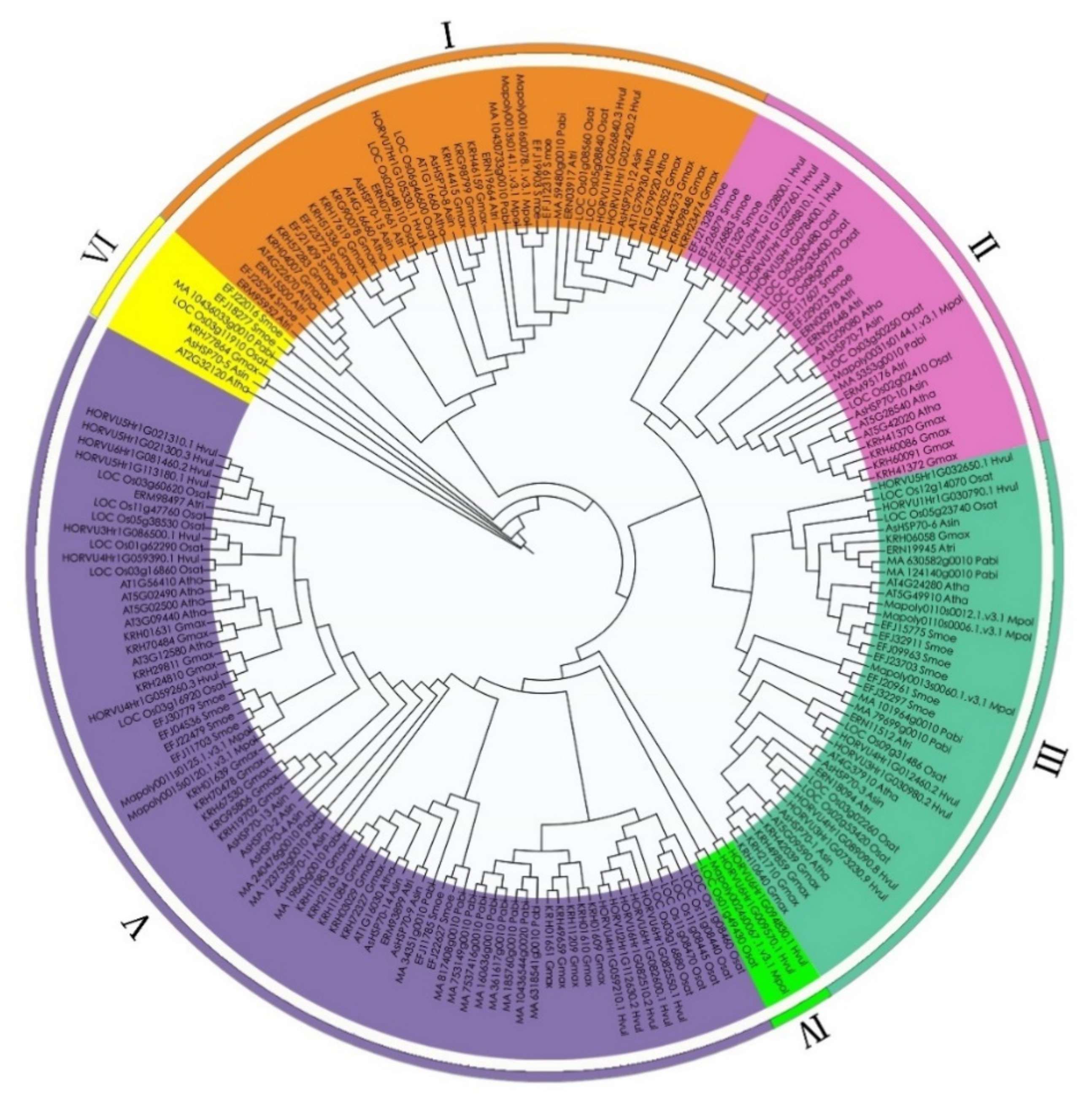
To evaluate the phylogenetic relationships among the AsHSP70 genes in A. sinensis and other plant species in depth, a comprehensive phylogenetic tree of HSP70 proteins from eight different plant species was performed by generating a neighbor-joining phylogenetic tree, including 15 A. sinensis HSP70s, 9 M. polymorpha HSP70s, 25 S. moellendorffii HSP70s, 19 A. trichopoda HSP70s, 41 G. max HSP70s, 13 A. thaliana HSP70s, 27 O. sativa HSP70s, 20 P. abies HSP70s and 27 H. vulgare HSP70s (Table 2). According to previous studies, the number of sub-families in O. sativa, G. max, A. thaliana are 6, 8, 5, respectively [22,25,43]. Based on their phylogenetic relationships, 196 HSP70 proteins were separated into the following six major sub-families: I, II, III, IV, V and VI (Figure 3). Among these six groups, except class VI and IV, all the other groups included nine plant species HSP70 members, as follows: class IV was the smallest sub-family, containing 2 H. vulgare, 1 O. sativa and 1 M. polymorpha; class VI contained 7 members, 2 S. moellendorffii, 1 from 5 plant species (P. abies, O. sativa, A. trichopoda, G. max and A. sinensis), and no H. vulgare, liverwort and A. thaliana. Cluster V was the largest group containing 76 members, 19 members from G. max, 6 from A. trichopoda, 2 from A. thaliana, 6 from S. moellendorffii, 11 from O. sativa, 12 from H. vulgare, 12 from P. abies, 2 from M. polymorpha and 6 from A. sinensis. Class I contained 41 members was the second largest with 3 A. sinensis, 5 A. thaliana, 5 S. moellendorffii, 5 A. trichopoda, 12 G. max, 4 O. sativa, 3 H. vulgare, 2 P. abies and 2 M. polymorpha. Class III was composed of 39 members (5 G. max, 3 A. sinensis, 3 A. thaliana, 4 A. trichopoda, 4 P. abies, 6 S. moellendorffii, 6 H. vulgare, 5 O. sativa and 3 M. polymorpha). The HSP70 numbers in class II were 4 G. max, 2 A. sinensis, 3 A. thaliana, 3 A. trichopoda, 1 P. abies, 6 S. moellendorffii, 4 H. vulgare, 5 O. sativa and 1 M. polymorpha.

Table 2.
HSP70 family members in selected plant species.
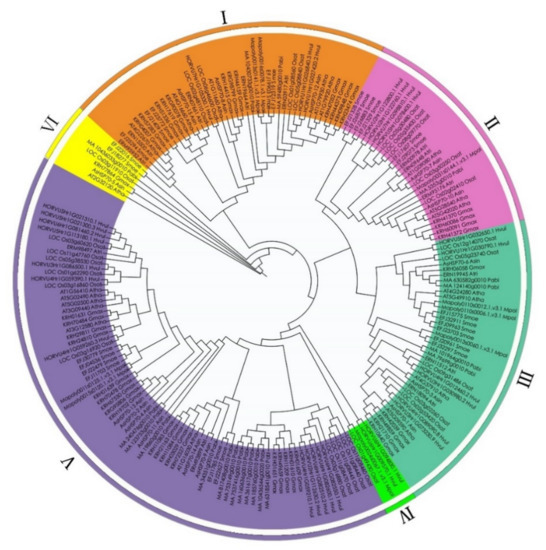
Figure 3.
Phylogenetic relationship of HSP70 in A. sin (Aquilaria sinensis), A. tha (Arabidopsis thaliana), A. tri (Amborella trichopoda), G. max (Glycine max), H. vul (Hordeum vulgare), M. pol (Marchantia polymorpha), O. sta (Oryza sativa), P. abies (Picea abies), S. moe (Selaginella moellendorffii). Based on the total identified 196 HSP70 homologs in 9 plant species, an unrooted phylogenetic tree was calculated with the maximum likelihood method, using JTT modeling with γ-distributed rates and 1000 bootstrap replications.
3.3. Conserved Motif and Gene Structure Analyses of HSP70 Genes in A. sinensis
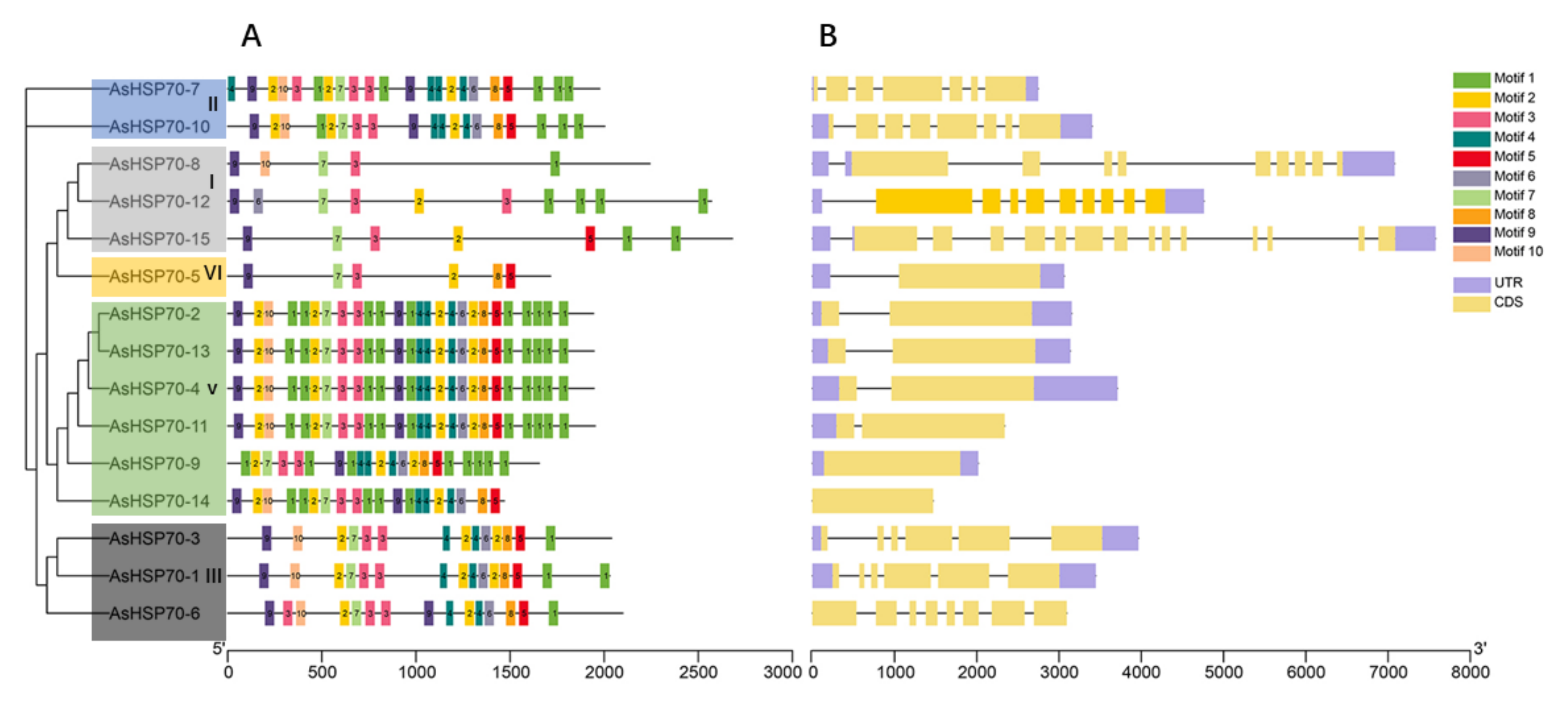
To better understand the evolution conservation of AsHSP70 family, the number and distribution of conserved motifs were identified using the MEME program. A total of 10 consensus motifs were detected based on domain compositions of HSP70-conserved amino acid compositions of identified motifs, as shown in Table 3. The results showed that most of the closely related members share similar motif patterns, suggesting possible functional similarity among these AsHSP70 proteins (Figure 4A). For all the 15 AsHSP70 proteins, four motifs (motif 1, motif 3, motif 7, motif 9) existed in all AsHSP70 proteins and motif 2 existed in almost all of them except AsHSP70-8. Except for the lack of motif 10 in AsHSP70-9, proteins from sub-family II, V and III contained all 10 conserved motifs. Over half of AsHSP70 were found to contain more than 19 motifs, while the proteins from sub-family I and VI, including AsHSP70-8, AsHSP70-2, AsHSP70-15 and AsHSP70-5, contained less than 10 motifs. Protein motifs from sub-family V were the most conserved, except AsHSP70-9, due to the fluctuant number and position of motifs. The type, number and position of conserved motifs are revealed be similar in the proteins from the same sub-family.

Table 3.
HSP70 motif sequences identified in A. sinensis by MEME tools.
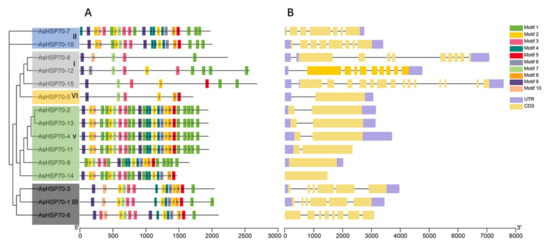
Figure 4.
Distribution of conserved motifs and gene structure of the AsHSP70. (A) Conserved motifs analysis of AsHSP70 proteins using MEME tools. Conserved motifs are shown in different colored boxes. (B) Exon–intron organization of AsHSP70 genes. Yellow boxes represent exons and black lines represent introns. The untranslated regions (UTRs) are indicated by purple boxes.
The genes’ exon–intron structure can reflect the evolution of gene families [44]. To better understand the functional diversification of the AsHSP70 proteins during their evolution, we further investigated the gene structure of 15 AsHSP70 genes. The results showed that the number of introns within ORFs varied among AsHSP70 genes, whereas the difference between genes of the same sub-family was small. Genes that belonged in the same cluster showed similar patterns, suggesting these AsHSP70 proteins contained similar function. (Figure 4B). The number of introns ranged from 0 to 14, and more than half of AsHSP70 genes contain five to nine introns. All members in sub-family V and VI contained one intron, with the exception that no intron was found in two AsHSP70 genes (AsHSP70-9 and AsHSP70-14). Genes from sub-family I (AsHSP70-8, AsHSP70-12, AsHSP70-15) contained higher numbers of intron than others. AsHSP70-15 was found to possess the largest number (14) of introns. As for sub-family II and III, the number of introns ranged from five to seven.
3.4. Expression Profiles of AsHSP70 Genes in Response to Heat Stress
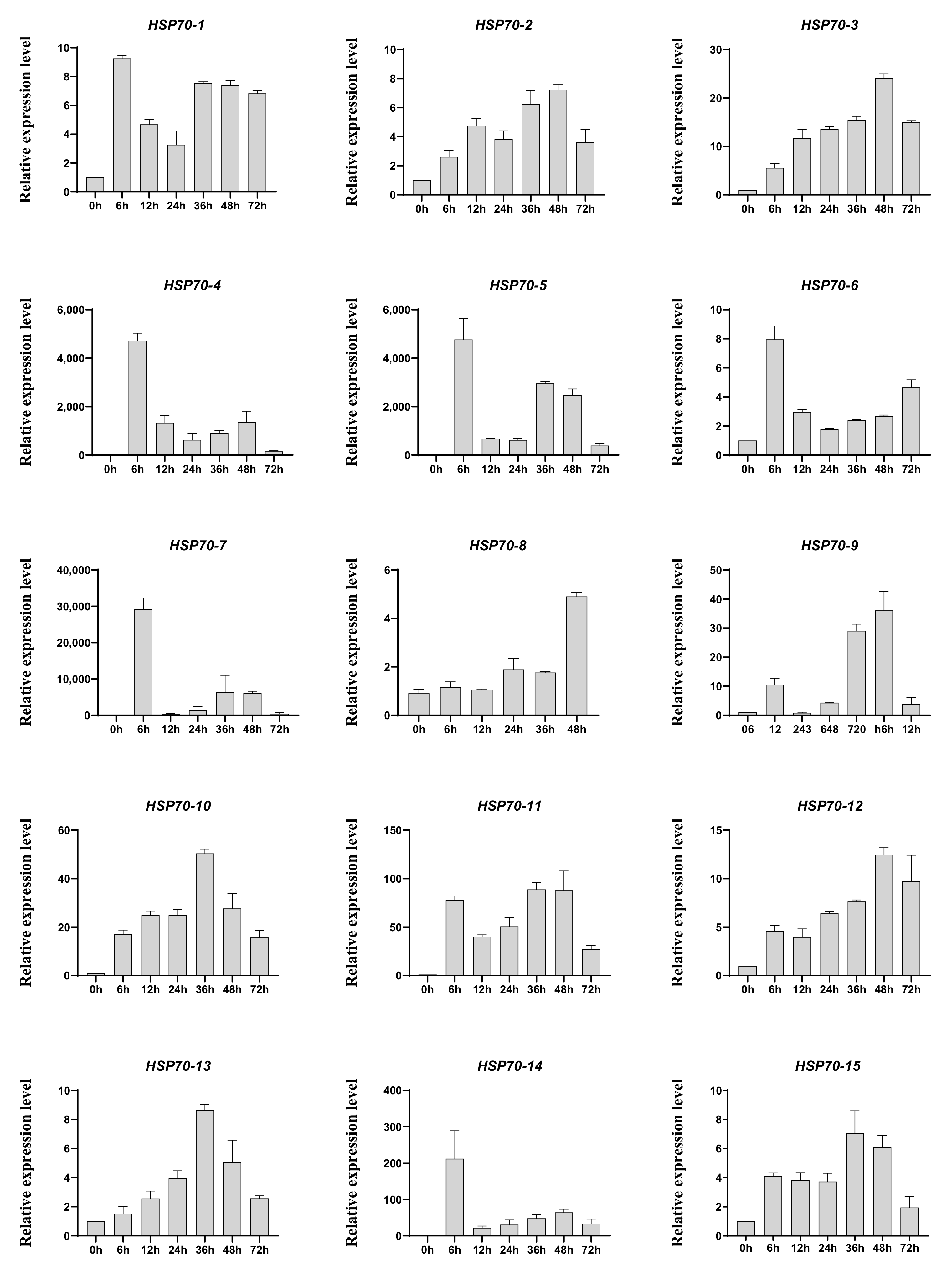
Heat shock proteins are a kind of protein that are expressed in large quantities after organisms are stimulated by stress, such as high temperature [45], and, among them, HSP70s play vital roles in response to heat treatment. In addition, the existing studies have shown that heat treatment is one of the traditional artificial methods to trigger agarwood formation [46], and our group have reported that heat shock can induce jasmonic acid production and the accumulation of agarwood sesquiterpene in A. sinensis cell suspension cultures [47]. Therefore, we first detected the response of AsHSP70s to heat shock treatment. The RNA was isolated from the heat stress-treated calli from different time points (0 h, 6 h, 12 h, 24 h, 36 h, 48 h, 72 h) and the relative expression profiles of 15 AsHSP70 genes were examined by qRT-PCR. As shown in Figure 5, the relative expression level of all 15 AsHSP70 genes were up-regulated by heat stress. The transcriptional levels of eight genes (AsHSP70-2, AsHSP70-3, AsHSP70-9, AsHSP70-10, AsHSP70-11, AsHSP70-12, AsHSP70-13 and AsHSP70-15) share similar expression patterns, they all increased in various degrees and reached the highest levels at 36 h or 48 h after heat stress treatment. The other six genes (AsHSP70-1, AsHSP70-4, AsHSP70-5, AsHSP70-6, AsHSP70-7 and AsHSP70-14) exhibited enhanced expression immediately after heat stress, with slowly or quickly decreased in the later period. Notably, among them, the expression level of three genes (AsHSP70-4, AsHSP70-5 and AsHSP70-7) was dramatically up-regulated (more than 1000-fold) compared with the control in response to heat stress. In contrast, AsHSP70-8 exhibited special expression patterns, its expression did not change and remained stable until 48 h after heat stress, with the following treatment, the expression level of AsHSP70-8 increased rapidly and peaked at 72 h.
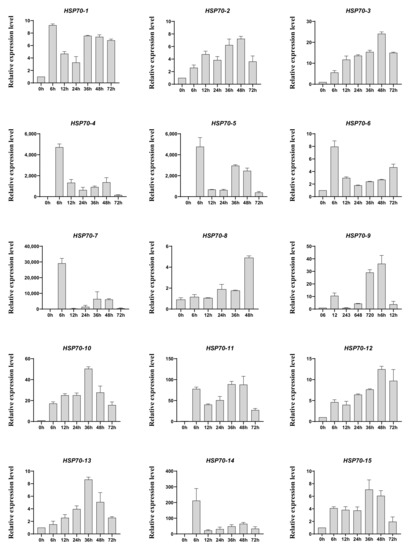
Figure 5.
Expression profiles of AsHSP70s in heat treated calli. A. sinensis calli were transferred from 25 °C to 42 °C and sampled at appointed times (0 h, 6 h, 12 h, 24 h, 36 h, 48 h, 72 h). Expression levels of all detected genes were assayed using real-time PCR analysis and AsGADPH as the internal control. Each value is the mean ± SE of 3 independent biological replicates.
3.5. Subcellular Localization of AsHSP70s
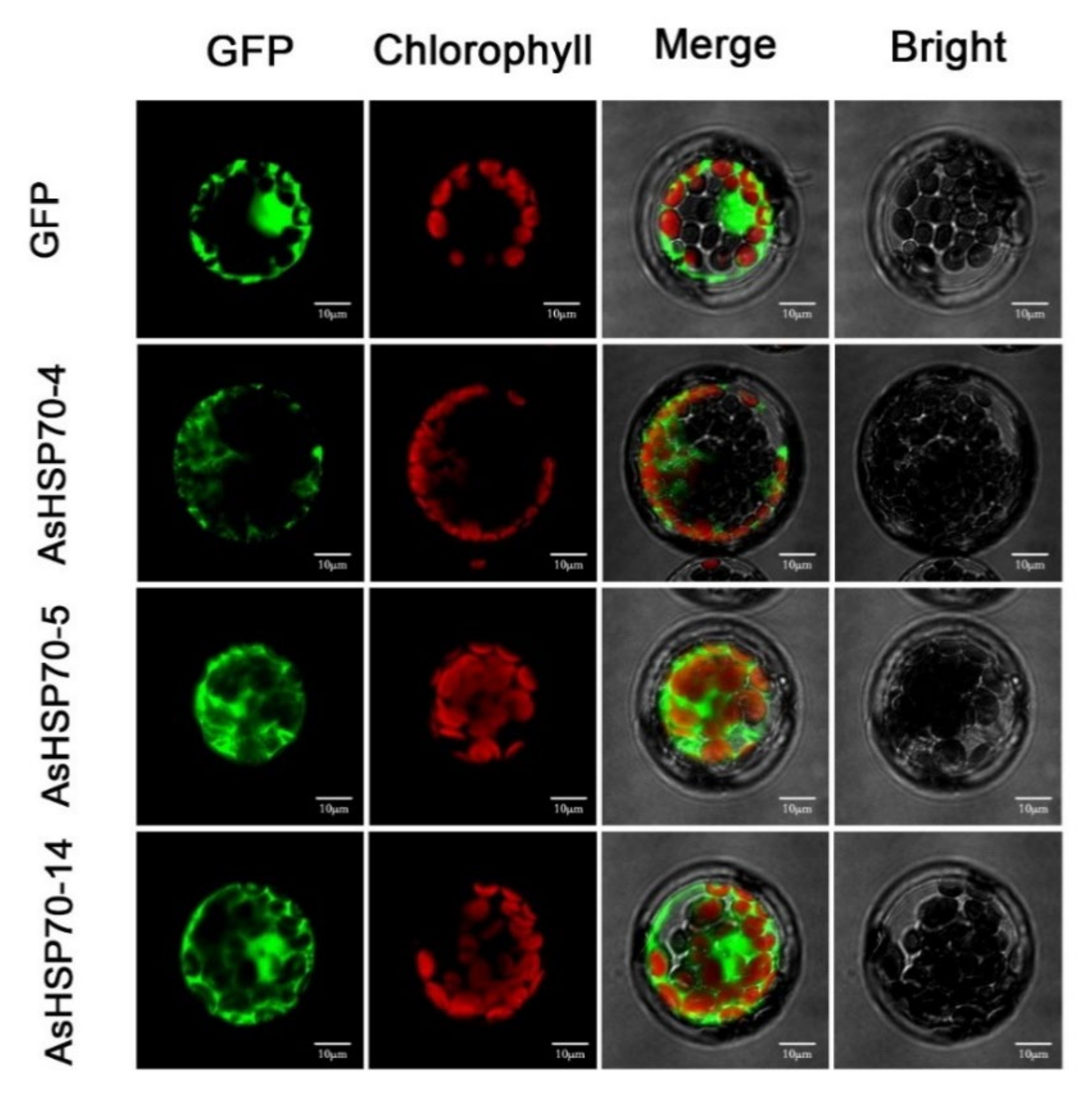
The protein subcellular localization prediction software WoLF PSORT was used to predict the protein localization of 15 candidate AsHSP70 in A. sinensis (Table 1). AsHSP70-10 and AsHSP70-15 were predicted to be localized in the ER. AsHSP70-1 and AsHSP70-3 were predicted to be localized in the mitochondria or in the chloroplast with high reliability, while AsHSP70-4 and AsHSP70-8 were likely localized in vacuole and nucleus, respectively. AsHSP70-6 was predicated to be localized in the chloroplast. For the other eight AsHSP70 proteins (AsHSP70-2, AsHSP70-5, AsHSP70-7, AsHSP70-9, AsHSP70-11, AsHSP70-12, AsHSP70-13 and AsHSP70-14), the cytosol is predicted to be their most likely location. To further confirm their predicted localizations, three genes (AsHSP70-4, AsHSP70-5 and AsHSP70-14) which are up-regulated strongly by heat treatment, were selected for subcellular localization by pBWA(V)HS-GFP translational fusion construct. The recombinant fusion was transiently expressed in Arabidopsis leaf protoplast. pBWA(V)HS GFP vector was transformed as the control and was detected in the nucleus and cytoplasm (Figure 6). As shown in Figure 6, the green fluorescence signals from AsHSP70-5 and AsHSP70-14 fusion proteins were both detected in the nuclei and cytoplasm. The prediction results of AsHSP70-5 were consistent with the experimental results, while AsHSP70-14 was not predicated to be localized in the nuclei. AsHSP70-4 fusion protein was localized in the cytoplasm, which was different from the predicted results.
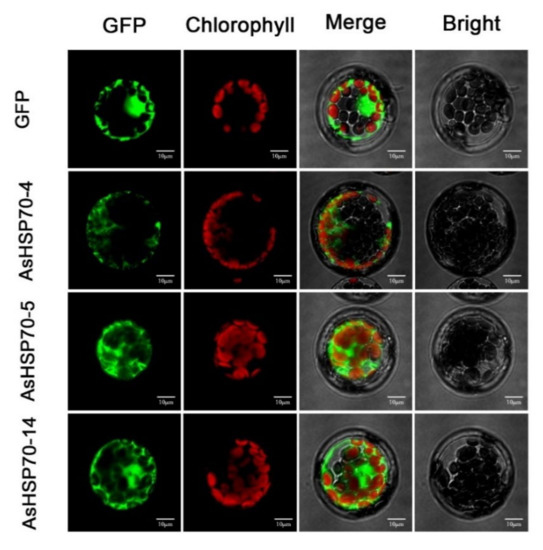
Figure 6.
Subcellular localization of AsHSP70 proteins in Arabidopsis protoplast. The AsHSP70-4-pBWA(V)HS-GFP, HSP70-5-pBWA(V)HS-GFP, HSP70-14-pBWA(V)HS-GFP and pBWA(V)HS-GFP vectors were transiently expressed in Arabidopsis protoplast. Confocal images were acquired at 48 h. Scale bar = 10 μm.
3.6. Expression Profiles of AsHSP70 in Different Tissues and Different Layers during the Whole-Tree-Inducing Agarwood Formation Process in A. sinensis
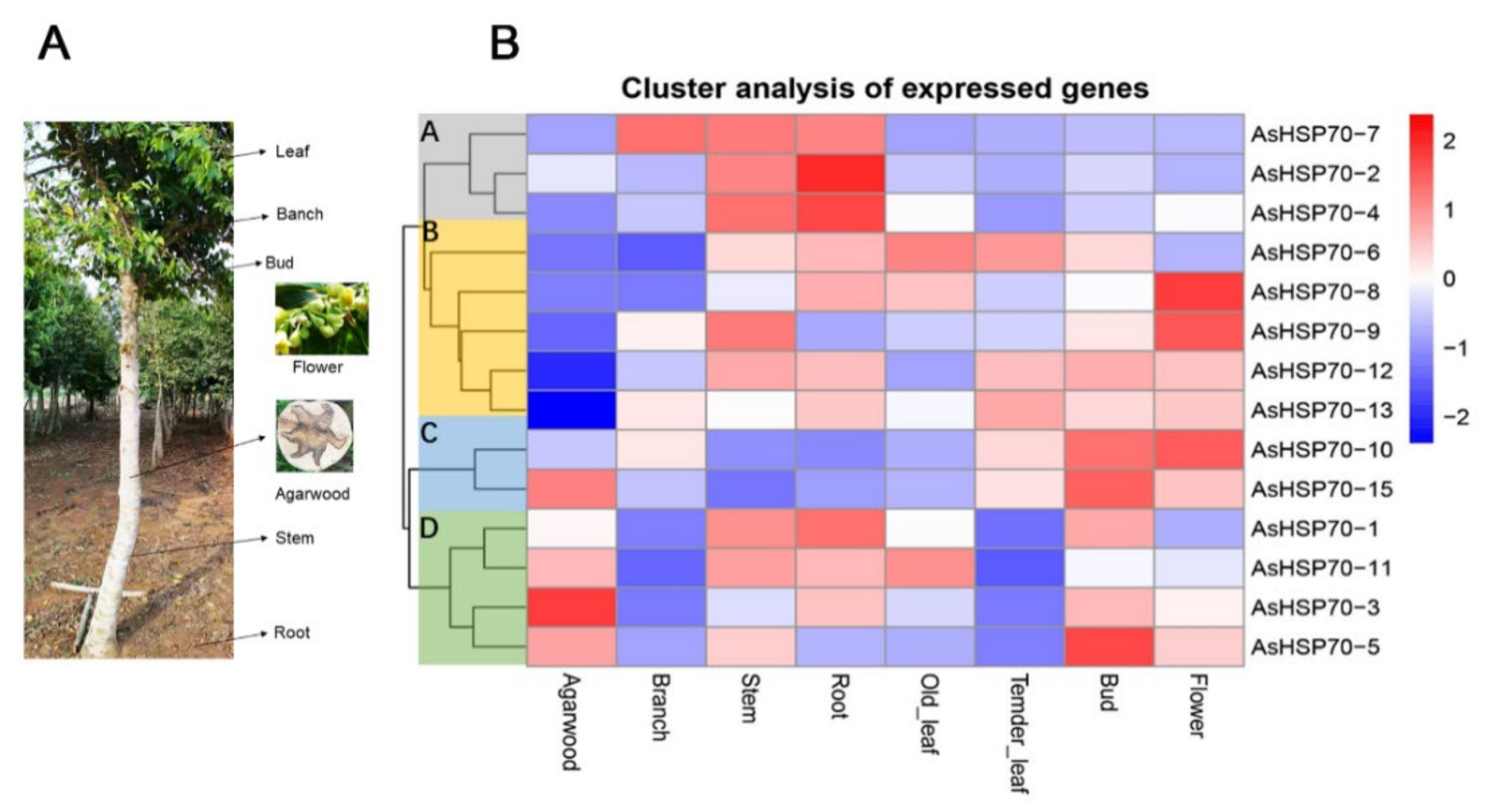
To investigate the temporal and spatial expression profile of the AsHSP70 genes in different tissues, expression levels of all 15 putative AsHSP70s were analyzed based on transcriptome data in 7 tissues, including agarwood, branch, stem, root, old leaves, tender leaves, bud and flower (Figure 7). The results revealed that all AsHSP70s genes exhibited broad expression patterns across various tissues, with the exception of the two AsHSP70s genes, AsHSP70-9 and AsHSP70-7. AsHSP70-9 that did not express in agarwood and AsHSP70-7 that did not express in agarwood and old leaves. The other 13 AsHSP70s genes were expressed in all 7 tissues. These findings indicate that AsHSP70s genes had diverse function and participated in multiple processes in A. sinensis. As a whole, AsHSP70s genes showed the highest expression in stem and the lowest expression in tender leaves. As shown in Figure 7A, 15 AsHSP70s genes were mainly clustered into four groups (A, B, C, D) according to their expression patterns, which presumes that they have similar function in the same cluster. In cluster A, all genes (AsHSP70-2, AsHSP70-4 and AsHSP70-7) exhibited high expression in stem and root, and AsHSP70-7 also showed high expression in branch. In cluster B and C, seven AsHSP70s genes (AsHSP70-6, AsHSP70-8, AsHSP70-9, AsHSP70-12, AsHSP70-13, AsHSP70-10, AsHSP70-15) were highly expressed in bud and flower, implying these AsHSP70s contained functions in vegetative organs. In cluster D, four AsHSP70s genes (AsHSP70-1, AsHSP70-11, AsHSP70-3, AsHSP70-5) showed high expression levels in agarwood, as well as AsHSP70-15. In contrast, the other three groups were the opposite, suggesting these five AsHSP70s genes may be closely related to the formation of agarwood.
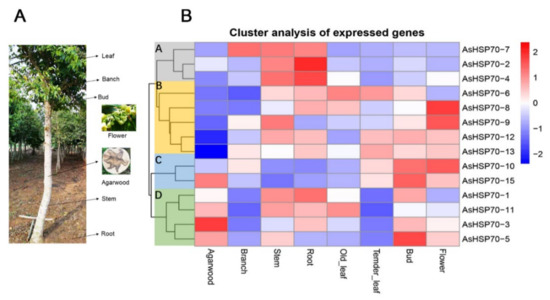
Figure 7.
Heat map of the AsHSP70 genes expression profile in different tissues and different tissues of A. sinensis trees. (A). The different tissues of A. sinensis trees. The agarwood layer was obtained after trees was wounded by external stimulus. (B). All gene expression levels were transformed to scores ranging from −2 to 2 and were colored blue, white, or red to represent low, moderate, or high expression levels, respectively.
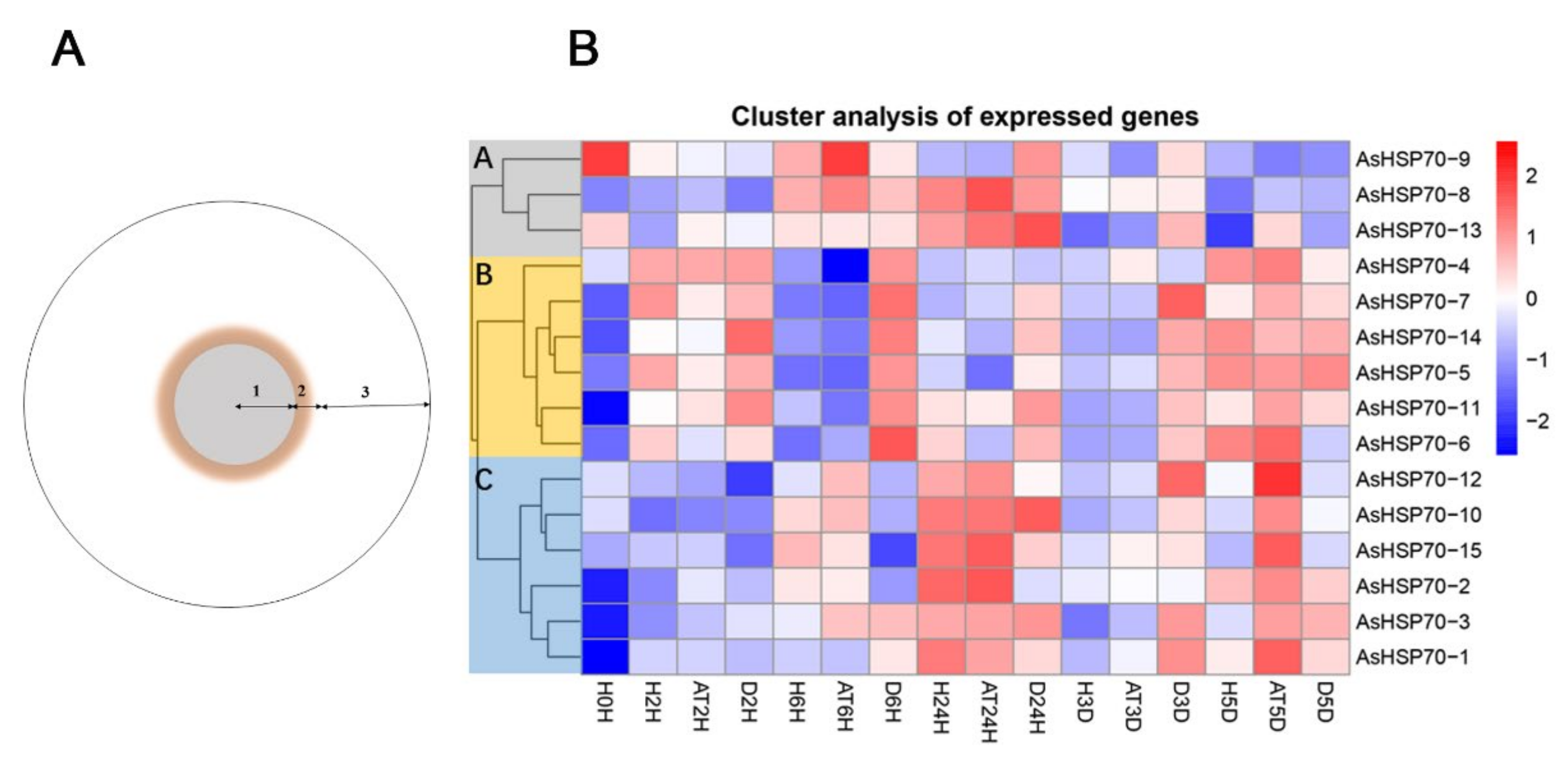
To evaluate whether AsHSP70 genes are involved in the progress of agarwood formation, transcriptome data were obtained from Agar-Wit inducing materials with different treatment time and different layers, the detail sections of agarwood are shown in Figure 8B. As shown in Figure 8A, all AsHSP70s were up-regulated in some layers or at certain time, suggesting all of them were involved in agarwood formation and play different functions in the process. In addition, 15 AsHSP70s genes were mainly clustered into three groups (A, B, and C) according to their expression patterns. Genes in group B were sensitive to the wound treatment and expressed mainly in decayed layer in early stage (from 2 h to 3 days), suggesting they contribute to cell death in the process of agarwood formation. These results are also consistent with the relative expression treated by heat, whereby genes in group B all exhibited enhanced expression immediately after heat stress. Among them, three genes (AsHSP70-4, AsHSP70-5, AsHSP70-7) expressed by a factor of over 103 after heat treatment. Genes in group C were mainly expressed in the agarwood layer, implying they probably have critical roles in agarwood formation, and their expression induced by heat shared similar patterns as well. Interestingly, during the Agar-Wit treatment, AsHSP70-8 from group A showed up-regulated expression from 6 h to 3 days, whereas the rest of the time they were downregulated.
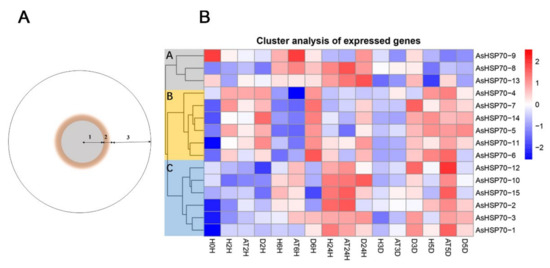
Figure 8.
Heat map of the AsHSP70 genes expression profile in different times under Agar- Wit treatment and different layers of agarwood in cross section of A. sinensis tree. (A) 1. the decayed layer after Agar-Wit treatment (D); 2. the agarwood and transition layer (AT); 3. the healthy layer (H). (B) All gene expression levels were transformed to scores ranging from −2 to 2 and were colored blue, white or red to represent low, moderate, or high expression levels, respectively.
4. Discussion
A. sinensis is a classic wound-induced medicinal plant and only can form agarwood after being wounded or infected by microbes [35,48]. The molecular mechanism of agarwood formation is still largely unclear. HSP70 is one of the most conserved proteins involved in the process of plant stress tolerance. To verify the functions of AsHSP70s in A. sinensis in response to stress, we performed a whole-genome analysis of the AsHSP70 gene family in A. sinensis for the first time. Overall, 15 AsHSP70 genes were identified in A. sinensis, which is less than other reported plant species (16 in C. quinoa, 18 in Arabidopsis, 61 in soybean) [22,23,25]. As shown in Figure 2, the majority of AsHSP70 genes were unevenly distributed on six chromosomes, mainly on both ends of the chromosome, a pattern that is consistent with that reported in Nicotiana tabacum and A. thaliana [22,43].
AsHSP70 can be divided into six sub-families in A. sinensis. Based on the phylogenetic tree, it was found that proteins from the same sub-family contained a similar type, order, and number of motifs, while these conditions were not seen among different sub-families. In the gene structure analysis, the same rule was also revealed, in that the most closely related AsHSP70 genes shared a more similar gene structure, while the same results were also found in a number of reported plants [49,50]. Gene structures revealed phylogenetic relationships and related to gene function [51]. The number of introns is usually related to the transcription regulation. Genes in group V and VI contained fewer introns, longer exons and more motifs, while genes in group I contained more introns and fewer motifs. This accords with earlier studies in other plants [43,52]. Genes with fewer introns are able to rapidly activate in timely response to various stresses; plants with a lower number of introns contain a stronger ability to adapt to external stimulus [53,54]. Combined with Figure 4 and Figure 5, genes in sub-family V and VI were highly induced after heat treatment, which supports the standpoints.
Heat treatment is used as an artificial method for producing high-quality agarwood and it can induce jasmonic acid production and the accumulation of agarwood sesquiterpene [47]. In this study, the expression profiles of AsHSP70 genes under heat treatment revealed that AsHSP70 genes were involved in responding to heat stress. As results showed, all AsHSP70 genes were up-regulated in heat stress, some AsHSP70 genes were induced at early stage, while some of them were induced later, indicating that different AsHSP70 genes played different roles during heat treatment, which was consistent with the results of expression profiles of different layers and time points treated with Agar-Wit.
HSP70 have been discovered in diverse subcellular compartments, such as the nuclei [4], mitochondrion [15] and endoplasmic reticulum [55]. In this study, AsHSP70 members were predicted located in various cellular compartments in A. sinensis. Over half of AsHSP70 proteins were located in cytoplasm or nuclei, similar results were also found in soybean in which more than half of HSP70 genes were localized in cytosol [25], according with the reports that cytosolic HSP proteins relocate to nuclei to protect cells from heat-induced nuclear damage [56]. In addition, with the analysis of subcellular localization of three AsHSP70 proteins (AsHSP70-5, AsHSP70-4 and AsHSP70-14), all located in the cytosol, two of these proteins (AsHSP70-5, AsHSP70-14) were also located in the nuclei. These results suggest HSP70s are preferred in specific cellular compartments, and AsHSP70s is involved in a defensive reaction with multiple organelles.
As shown in Figure 7, AsHSP70 genes were expressed in different tissues and organs in A. sinensis, indicating that they might participate in various biological processes of plants. Five genes (AsHSP70-1/3/5/11/15) were highly expressed in agarwood layers, implying they were promoters of agarwood formation. Combined with the expression patterns of different layers and time points during the agarwood formation (Figure 8A), AsHSP70-5 and AsHSP70-11 were expressed mainly in the decayed layer from 2 h to 3 days, then expressed in the agarwood layer at 5 days, while AsHSP70-1, AsHSP70-3 and AsHSP70-15 mainly expressed in the agarwood layer at 24 h. These results are consistent with their expression patterns induced by heat shock. In addition, we further explore the potential role of AsHSP70 in agarwood formation through the expression profiles in the process of agarwood formation treated with Agar-Wit. As shown in Figure 8A, genes in group B (AsHSP70-4, AsHSP70-5, AsHSP70-6, AsHSP70-7, AsHSP70-11 and AsHSP70-14) expressed specifically in the decayed layer at early stage (from 2 h to 24 h), implying they may play a regulatory role in cell death in the process of agarwood formation. Heat shock proteins help plants recover from stress through repairing damaged proteins or degrading them, thus restoring protein homeostasis and promoting cell survival. The results of qRT-PCR also showed that they were all induced immediately. Genes in group C (AsHSP70-1, AsHSP70-2, AsHSP70-3, AsHSP70-10, AsHSP70-12 and AsHSP70-15) expressed highly in the agarwood layer at 24 h, and all of them showed similar expression patterns after heat treatment, suggesting they might play a positive regulator in producing agarwood. These results suggest that the regulatory mechanism of agarwood formation is complicated, and that different kinds of AsHSP70 proteins play various function in different stages of the agarwood formation process.
5. Conclusions
In this study, we performed a comprehensive analysis of the HSP70 genes in A. sinensis covering phylogeny, chromosomal location, conserved motifs, gene structures, expression profiles and subcellular localization. In total, 15 AsHSP70 genes were identified and classed into six groups. The motifs and gene structures within the same subfamilies shared a similar pattern. The qRT-PCR results showed that the expression pattern of 15 AsHSP70 genes were all affected by heat stresses. Combined with the expression profiles of different tissues, and expression patterns in the different layers after the Agar-Wit treatment, different AsHSP70 genes are involved in different stages of formatting agarwood and contribute to various biological functions. This study lays a solid foundation for further functional analyses of AsHSP70 genes in A. sinensis.
Author Contributions
Y.X. and J.W. conceived and designed the experiments. C.Y., M.R. and Y.L. performed the experiments. C.Y. and P.S. analyzed the data; C.Y., Y.X. and J.W. wrote the paper. C.Y., Y.L., Y.X. and J.W. revised the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2018YFC1706400), National Natural Science Foundation of China (82073967, 82173925, 82104332), China Agriculture Research System of MOF and MARA, and CAMS Innovation Fund for Medical Sciences (2021-1-I2M-032).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, W.-X.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Wang, W.-X.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genom. 2007, 8, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.K.; Choi, Y.J. A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol. Lett. 2008, 31, 597–606. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-M.; Rhee, J.-S.; Jeong, C.-B.; Seo, J.S.; Park, G.S.; Lee, Y.-M.; Lee, J.-S. Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2014, 166, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Heckathorn, S.A.; Mueller, J.K.; LaGuidice, S.; Zhu, B.; Barrett, T.; Blair, B.; Dong, Y. Chloroplast small heat-shock proteins protect photosynthesis during heavy metal stress. Am. J. Bot. 2004, 91, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Neumann, D.; Lichtenberger, O.; Tschiersch, K.; Nover, L. Heat-shock proteins induce heavy-metal tolerance in higher plants. Planta 1994, 194, 360–367. [Google Scholar] [CrossRef]
- Vierling, E. The Roles of Heat Shock Proteins in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragovic, Z.; Broadley, S.A.; Shomura, Y.; Bracher, A.; Hartl, F.U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006, 25, 2519–2528. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural Analysis of Substrate Binding by the Molecular Chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [Green Version]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nat. Cell Biol. 1990, 346, 623–628. [Google Scholar] [CrossRef]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [Green Version]
- Alvim, F.C.; Carolino, S.M.B.; Cascardo, J.C.M.; Nunes, C.C.; Martinez, C.A.; Otoni, W.C.; Fontes, E.P.B. Enhanced Accumulation of BiP in Transgenic Plants Confers Tolerance to Water Stress. Plant Physiol. 2001, 126, 1042–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.K.; Hong, C.B. Over-expression of tobacco NtHSP70-1 contributes to drought-stress tolerance in plants. Plant Cell Rep. 2006, 25, 349–358. [Google Scholar] [CrossRef]
- Augustine, S.M.; Cherian, A.V.; Syamaladevi, D.P.; Subramonian, N. Erianthus arundinaceus HSP70 (EaHSP70) acts as a key regulator in the formation of anisotropic interdigitation in sugarcane (Saccharum spp. hybrid) in response to drought stress. Plant Cell Physiol. 2015, 56, 2368–2380. [Google Scholar] [CrossRef] [Green Version]
- Mosaviazam, B.; Ramezani, A.; Morowvat, M.H.; Niazi, A.; Mousavi, P.; Moghadam, A.; Zarrini, G.; Ghasemi, Y. HSP70 Gene Expression Analysis in Dunaliella salina Under Salt Stress. Int. J. Pharm. Phytochem. Res. 2016, 8, 767–770. [Google Scholar]
- Ndimba, B.K.; Thomas, L.A.; Ngara, R. Sorghum 2-Dimensional Proteome Profiles and Analysis of HSP70 Expression Under Salinity Stress. Agric. Nat. Resour. 2010, 44, 768–775. [Google Scholar]
- Su, P.-H.; Li, H.-M. Arabidopsis Stromal 70-kD Heat Shock Proteins Are Essential for Plant Development and Important for Thermotolerance of Germinating Seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latijnhouwers, M.; Xu, X.-M.; Møller, S.G. Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 2010, 232, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.-L.; Wang, J.-S.; Barakat, A.; Liu, H.-C.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 201. [Google Scholar] [CrossRef]
- Liu, J.; Wang, R.; Liu, W.; Zhang, H.; Guo, Y.; Wen, R. Genome-Wide Characterization of Heat-Shock Protein 70s from Chenopodium quinoa and Expression Analyses of Cqhsp70s in Response to Drought Stress. Genes 2018, 9, 35. [Google Scholar] [CrossRef] [Green Version]
- Divya, K.; Kishor, P.B.K.; Singam, P.; Maraka, N.; Vadez, V.; Reddy, P.S.; Bhatnagar-Mathur, P. Genome-wide Identification and Characterization of Hsp70 gene family in Pearl millet (Pennisetumglaucum). Curr. Trends Biotechnol. Pharm. 2019, 13, 102–111. [Google Scholar]
- Zhang, L.; Zhao, H.-K.; Dong, Q.-L.; Zhang, Y.-Y.; Wang, Y.-M.; Li, H.-Y.; Xing, G.-J.; Li, Q.-Y.; Dong, Y.-S. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.W.; Xu, Y.H.; Yu, C.C.; Lv, F.F.; Tang, X.L.; Gao, Z.H.; Zhang, Z.; Wang, H.; Liu, Y.; Wei, J.H. WRKY44 represses expression of the wound-induced sesquiterpene biosynthetic gene ASS1 in Aquilaria sinensis. J. Exp. Bot. 2020, 71, 1128–1138. [Google Scholar]
- Takemoto, H.; Ito, M.; Shiraki, T.; Yagura, T.; Honda, G. Sedative effects of vapor inhalation of agarwood oil and spikenard extract and identification of their active components. J. Nat. Med. 2007, 62, 41–46. [Google Scholar] [CrossRef]
- Kakino, M.; Tazawa, S.; Maruyama, H.; Tsuruma, K.; Araki, Y.; Shimazawa, M.; Hara, H. Laxative effects of agarwood on low-fiber diet-induced constipation in rats. BMC Complement. Altern. Med. 2010, 10, 68. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, C.; Espinoza, E. Evaluating agarwood products for 2-(2-phenylethyl)chromones using direct analysis in real time time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 2649–2656. [Google Scholar] [CrossRef]
- Mei, W.-L.; Yang, D.-L.; Wang, H.; Yang, J.-L.; Zeng, Y.-B.; Guo, Z.-K.; Dong, W.-H.; Li, W.; Dai, H.-F. Characterization and Determination of 2-(2-Phenylethyl)chromones in Agarwood by GC-MS. Molecules 2013, 18, 12324–12345. [Google Scholar] [CrossRef] [PubMed]
- China Pharmacopoeia Committee. The Pharmacopoeia of People’s Republic of China (I); Chemical Industry Press: Beijing, China, 2020. [Google Scholar]
- Zhang, Z.; Wei, J.; Han, X.; Liang, L.; Yang, Y.; Meng, H.; Xu, Y.; Gao, Z. The Sesquiterpene Biosynthesis and Vessel-Occlusion Formation in Stems of Aquilaria sinensis (Lour.) Gilg Trees Induced by Wounding Treatments without Variation of Microbial Communities. Int. J. Mol. Sci. 2014, 15, 23589–23603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, X.; Yang, Y.; Wei, J.-H.; Meng, H.; Gao, Z.-H.; Xu, Y.-H. Hydrogen peroxide induces vessel occlusions and stimulates sesquiterpenes accumulation in stems of Aquilaria sinensis. Plant Growth Regul. 2013, 72, 81–87. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, X.; Yang, Y.; Sui, C.; Xu, Y.; Wei, J. Interxylary phloem and xylem rays are the structural foundation of agarwood resin formation in the stems of Aquilaria sinensis. Trees 2018, 33, 533–542. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Z.; Wang, M.; Wei, J.; Chen, H.; Gao, Z.; Sui, C.; Luo, H.; Zhang, X.; Yang, Y.; et al. Identification of genes related to agarwood formation: Transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genom. 2013, 14, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Yang, Y.; Xue, J.; Wei, J.; Zhang, Z.; Chen, H. Comparison of Compositions and Antimicrobial Activities of Essential Oils from Chemically Stimulated Agarwood, Wild Agarwood and Healthy Aquilaria sinensis (Lour.) Gilg Trees. Molecules 2011, 16, 4884–4896. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucl. Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.H.; Yang, Y.; Zhang, Z.; Meng, H.; Feng, J.D.; Gan, B.C. Production of agarwood in Aquilaria sinensis trees via transfusion technique. China Patent CN101755629 B, 1 February 2010. [Google Scholar]
- Kim, J.; Somers, D.E. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 2010, 154, 611–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Pan, F.; Lou, X.; Wang, D.; Yang, C.; Zhang, B.; Zhang, H. Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum. Mol. Biol. Rep. 2019, 46, 1941–1954. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.L.; Goepfert, S.; Peitsch, M.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, P. Heat shock factors in rice (Oryza sativa L.): Genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genom. 2011, 286, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, H.; Yang, Y.; Zhang, Z.; Wei, J.; Meng, H.; Chen, W.; Feng, J.; Gan, B.; Chen, X.; et al. Whole-tree Agarwood-Inducing Technique: An Efficient Novel Technique for Producing High-Quality Agarwood in Cultivated Aquilaria sinensis Trees. Molecules 2013, 18, 3086–3106. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-H.; Liao, Y.-C.; Zhang, Z.; Liu, J.; Sun, P.-W.; Gao, Z.-H.; Sui, C.; Wei, J.-H. Jasmonic acid is a crucial signal transducer in heat shock induced sesquiterpene formation in Aquilaria sinensis. Sci. Rep. 2016, 6, 21843. [Google Scholar] [CrossRef] [PubMed]
- Tamuli, P.; Boruah, P.; Samanta, R. Biochemical changes in agarwood tree (Aquilaria malaccensis Lamk.) during pathogenesis. J. Spices Aromat. Crop. 2004, 13, 87–91. [Google Scholar]
- Su, H.; Xing, M.; Liu, X.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Wang, Y.; Lv, H. Genome-wide analysis of HSP70 family genes in cabbage (Brassica oleracea var. capitata) reveals their involvement in floral development. BMC Genom. 2019, 20, 369. [Google Scholar] [CrossRef]
- Liu, J.; Pang, X.; Cheng, Y.; Yin, Y.; Zhang, Q.; Su, W.; Hu, B.; Guo, Q.; Ha, S.; Zhang, J.; et al. The Hsp70 Gene Family in Solanum tuberosum: Genome-Wide Identification, Phylogeny, and Expression Patterns. Sci. Rep. 2018, 8, 16628. [Google Scholar] [CrossRef]
- Skolnick, J.; Fetrow, J. From genes to protein structure and function: Novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000, 18, 34–39. [Google Scholar] [CrossRef]
- Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Chandrasekaran, U.; Liu, A. Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genom. 2014, 36, 527–537. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kose, S.; Furuta, M.; Imamoto, N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell 2012, 149, 578–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).