Whole-Exome Sequencing Reveals Pathogenic SIRT1 Variant in Brain Arteriovenous Malformation: A Case Report
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. DNA Isolation
2.3. Whole Exome Sequencing (WES)
2.4. WES Data Processing
2.5. Gene Ontology (GO), Disease and Pathway Over-Representation Analysis (ORA)
2.6. Mutation Validation
3. Results
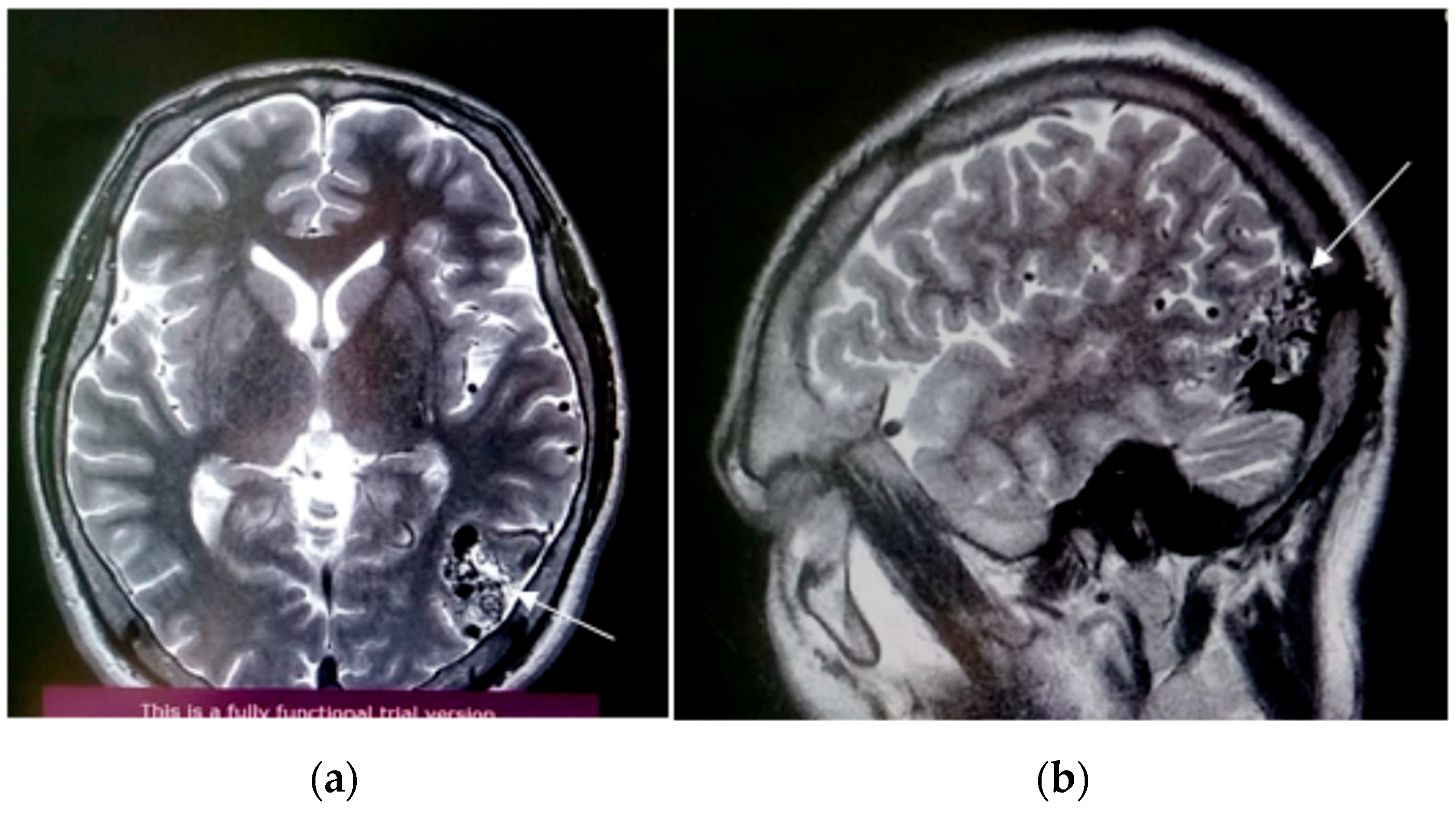
3.1. Case Presentation and Surgical Procedure
3.2. WES Results Analysis
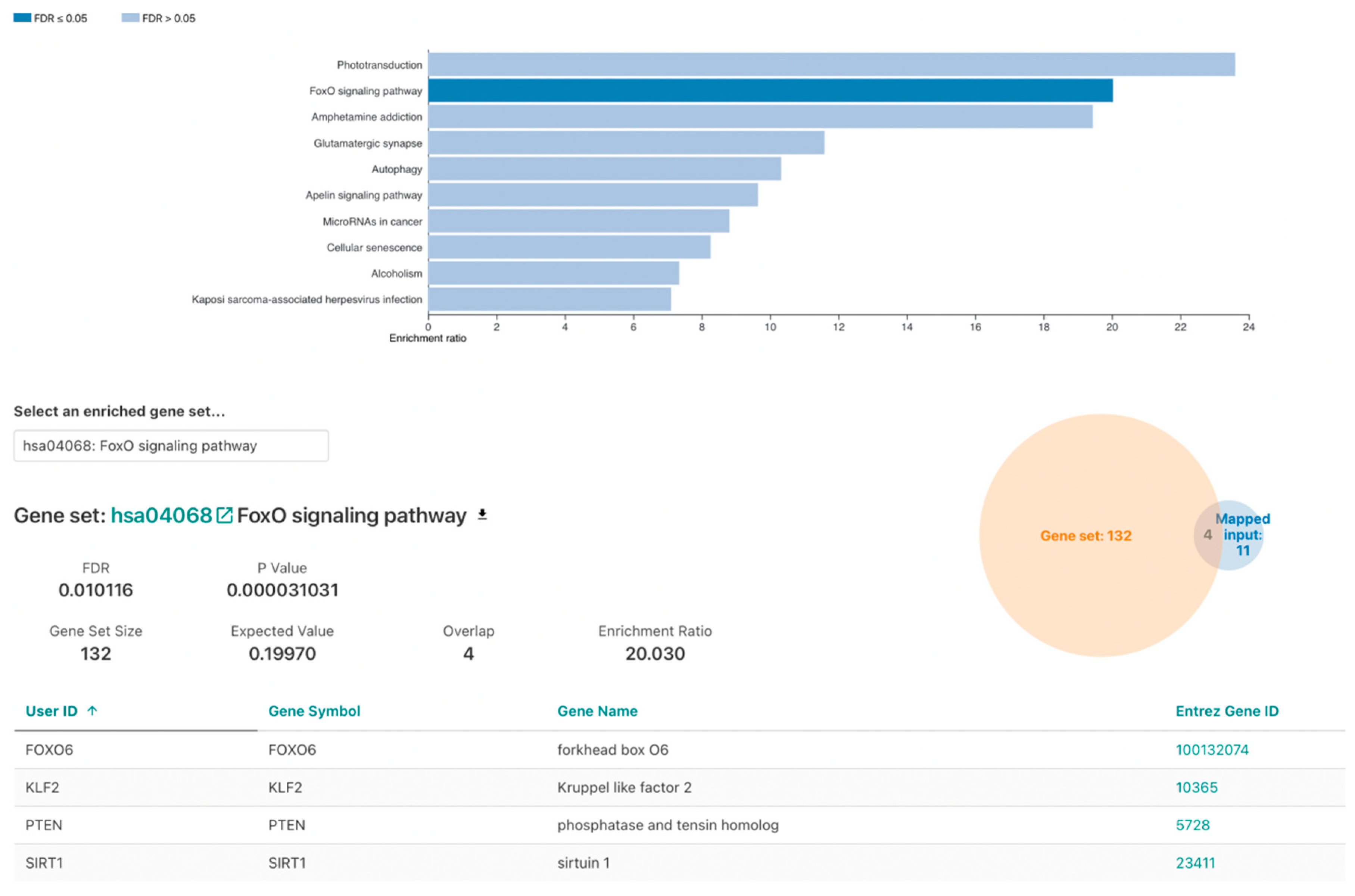
3.3. Over-Representation Analysis
3.4. FOXO Signaling Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Laakso, A.; Hernesniemi, J. Arteriovenous malformations: Epidemiology and clinical presentation. Neurosurg. Clin. N. Am. 2012, 23, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dalton, A.; Dobson, G.; Prasad, M.; Mukerji, N. De novo intracerebral arteriovenous malformations and a review of the theories of their formation. Br. J. Neurosurg. 2018, 32, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Bharatha, A.; Faughnan, M.E.; Kim, H.; Pourmohamad, T.; Krings, T.; Bayrak-Toydemir, P.; Pawlikowska, L.; McCulloch, C.E.; Lawton, M.T.; Dowd, C.F.; et al. Multiplicity of brain arteriovenous malformations predicts the diagnosis of hereditary hemorrhagic telangiectasia. Stroke 2012, 43, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, B.; Toktaş, Z.O.; Akakın, A.; Işık, S.; Bilguvar, K.; Kılıç, T.; Günel, M. Familial occurrence of brain arteriovenous malformation: A novel ACVRL1 mutation detected by whole exome sequencing. J. Neurosurg. 2017, 126, 1879–1883. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M. Pathogenesis of brain arteriovenous malformations. Neurol. Med. Chir. 2016, 56, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Parambil, J.G. Hereditary hemorrhagic telangiectasia. Clin. Chest Med. 2016, 37, 513–521. [Google Scholar] [CrossRef]
- Eerola, I.; Boon, L.M.; Mulliken, J.B.; Burrows, P.E.; Dompmartin, A.; Watanabe, S.; Vanwijck, R.; Vikkula, M. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am. J. Hum. Genet. 2003, 73, 1240–1249. [Google Scholar] [CrossRef]
- Pabaney, A.H.; Rammo, R.A.; Tahir, R.A.; Seyfried, D. Development of de novo arteriovenous malformation following ischemic stroke: Case report and review of current literature. World Neurosurg. 2016, 96, 608.e5–608.e12. [Google Scholar] [CrossRef]
- Weinsheimer, S.; Bendjilali, N.; Nelson, J.; Guo, D.E.; Zaroff, J.G.; Sidney, S.; McCulloch, C.E.; Al-Shahi Salman, R.; Berg, J.N.; Koeleman, B.P.; et al. Genome-wide association studies of sporadic brain arteriovenous malformations. J. Neurol. Neurosurg. Psychiatry 2016, 87, 916–923. [Google Scholar] [CrossRef]
- Thomas, J.M.; Surendran, S.; Abraham, M.; Rajavelu, A.; Chandrasekharan, C. Kartha genetic and epigenetic mechanisms in the development of arteriovenous malformations in the brain. Clin. Epigenet. 2016, 8, 78. [Google Scholar] [CrossRef]
- Nikolaev, S.I.; Vetiska, S.; Bonilla, X.; Boudreau, E.; Jauhiainen, S.; Rezai Jahromi, B.; Khyzha, N.; DiStefano, P.V.; Suutarinen, S.; Kiehl, T.R.; et al. Somatic activation of KRAS mutations in arteriovenous malformations in the brain. N. Engl. J. Med. 2018, 378, 250–261. [Google Scholar] [CrossRef]
- Hong, T.; Yan, Y.; Li, J.; Radovanovic, I.; Ma, X.; Shao, Y.W.; Yu, J.; Ma, Y.; Zhang, P.; Ling, F.; et al. High prevalence of KRAS/BRAF somatic mutations in brain and spinal cord arteriovenous malformation. Brain 2019, 142, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Walcott, B.P.; Winkler, E.A.; Zhou, S.; Birk, H.; Guo, D.; Koch, M.J.; Stapleton, C.J.; Spiegelman, D.; Dionne-Laporte, A.; Dion, P.A.; et al. Identification of a rare BMP pathway mutation in a non-syndromic human brain arteriovenous malformation using exome sequencing. Hum. Genome Var. 2018, 5, 18001. [Google Scholar] [CrossRef] [PubMed]
- Sauve, A.A.; Wolberger, C.; Schramm, V.L.; Boeke, J.D. Bbiochemistry of the sirtuins. Annu. Rev. Biochem. 2006, 75, 435–465. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and non-conserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar] [CrossRef]
- Cheng, H.L.; Mostoslavsky, R.; Saito, S.; Manis, J.P.; Gu, Y.; Patel, P.; Bronson, R.; Appella, E.; Alt, F.W.; Chua, K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 2003, 100, 10794–10799. [Google Scholar] [CrossRef]
- Nakagawa, T.; Guarente, L. Sirtuins. J. Cell Sci. 2011, 124, 833–838. [Google Scholar] [CrossRef]
- Man, A.W.; Li, H.; Xia, N. Role of Sirtuin1 in regulating endothelial function, arterial remodeling, and vascular aging/front physiology. Front. Physiol. 2019, 10, 1173. [Google Scholar] [CrossRef]
- Libert, S.; Pointer, K.; Bell, E.L.; Das, A.; Cohen, D.E.; Asara, J.M.; Kapur, K.; Bergmann, S.; Preisig, M.; Otowa, T.; et al. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell 2011, 147, 1459–1472. [Google Scholar] [CrossRef]
- Abuzenadah, A.; Al-Saedi, S.; Karim, S.; Al-Qahtani, M. Role of Overexpressed Transcription Factor FOXO1 in Fatal Cardiovascular Septal Defects in Patau Syndrome: Molecular and Therapeutic Strategies. Int. J. Mol. Sci. 2018, 19, 3547. [Google Scholar] [CrossRef]
- Dagra, A.; Williams, E.; Aghili-Mehrizi, S.; Goutnik, M.A.; Martinez, M.; Turner, R.C. Lucke-Wold B Pediatric Subarachnoid Hemorrhage: Rare Events with Important Implications. Brain Neurol. Disord. 2022, 5, 20. [Google Scholar] [CrossRef]
- Potente, M.; Ghaeni, L.; Baldessari, D.; Mostoslavsky, R.; Rossig, L.; Dequiedt, F.; Haendeler, J.; Mione, M.; Dejana, E.; Alt, F.W.; et al. SIRT1 controls endothelial angiogenic function during vascular growth. Genes Dev. 2007, 21, 2644–2658. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Ko, Y.S.; Park, J.; Choi, Y.; Park, J.-W.; Kim, Y.; Pyo, J.S.; Yoo, Y.B.; Lee, J.S.; Lee, B.L. Forkhead Transcription Factor FOXO1 Inhibits Angiogenesis in Gastric Cancer in Relation to SIRT1. Cancer Res. Treat. 2016, 48, 345–354. [Google Scholar] [CrossRef] [PubMed]


| AVM | Blood | |
|---|---|---|
| Capture method | Agilent SureSelect Human All Exon V6 | |
| Mean coverage | ||
| % > 10× | 99.0 | 98.8 |
| % > 20× | 97.0 | 96.3 |
| % > 30× | 93.6 | 92.1 |
| Primer | Sequence (5′–3′) | Ta (°C) | Amplicon Length (bp) |
|---|---|---|---|
| SIRT_F | GACCCGTAGTGTTGTGGTCT | 64 | 558 |
| SIRT_R | TCGTCTTCGTCGTACAAGTTGTC | 64 |
| N | Chr | Position | SNP | Gene | REF | ALT | Type | Blood | Tissue |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | 41847882 | rs373524987 | FOXO6 | CA | - | frameshift deletion | 0/1 | 1/1 |
| 2 | chr1 | 41847886 | FOXO6 | C | G | nonsynonymous SNV | 0/1 | 1/1 | |
| 3 | chr10 | 89623901 | rs2943772 | PTEN | G | C | nonsynonymous SNV | / | 1/1 |
| 4 | chr20 | 56803624 | rs146771462 | ANKRD60 | G | C | nonsynonymous SNV | 1/1 | 0/0 |
| 5 | chr3 | 75786555 | rs141276988 | ZNF717 | - | TG | frameshift insertion | 0/1 | 0/0 |
| 6 | chr3 | 112253058 | rs35560667 | ATG3 | - | A | frameshift insertion | 0/1 | 1/1 |
| 7 | chr1 | 1355796 | rs145378993 | ANKRD65 | C | T | nonsynonymous SNV | 0/1 | 0/0 |
| 8 | chr10 | 69644589 | rs548590752 | SIRT1 | C | T | nonsynonymous SNV | 0/0 | 0/1 |
| 9 | chr16 | 72042682 | rs3213422 | DHODH | A | C | nonsynonymous SNV | 0/1 | 1/1 |
| 10 | chr17 | 16256681 | rs188652843 | CENPV | G | A | nonsynonymous SNV | 1/1 | 0/1 |
| 11 | chr19 | 1000785 | rs12986002 | GRIN3B | C | T | nonsynonymous SNV | 1/1 | 0/1 |
| 12 | chr19 | 16436376 | rs3745319 | KLF2 | G | A | nonsynonymous SNV | 0/0 | 0/1 |
| 13 | chr2 | 26407937 | rs181971256 | GAREM2 | A | G | nonsynonymous SNV | 0/0 | 0/1 |
| 14 | chr2 | 100938226 | rs74177694 | LONRF2 | G | C | nonsynonymous SNV | / | 1/1 |
| 15 | chr2 | 100938481 | rs74177696 | LONRF2 | C | G | nonsynonymous SNV | 0/0 | 0/1 |
| 16 | chr2 | 128459214 | rs10206957 | SFT2D3 | C | G | nonsynonymous SNV | 1/1 | 0/1 |
| 17 | chr2 | 202410300 | rs10804117 | C2CD6 | A | T | nonsynonymous SNV | 0/0 | 0/1 |
| 18 | chr22 | 18923745 | rs2008720 | PRODH | G | T | nonsynonymous SNV | 0/0 | 0/1 |
| 19 | chr22 | 19137658 | rs34341950 | GSC2 | G | A | nonsynonymous SNV | 1/1 | 0/1 |
| 20 | chr3 | 75788076 | rs146581110 | ZNF717 | A | C | nonsynonymous SNV | / | 1/1 |
| 21 | chr3 | 75788085 | rs201605431 | ZNF717 | A | G | nonsynonymous SNV | / | 1/1 |
| 22 | chr3 | 75788105 | rs796745611 | ZNF717 | C | T | nonsynonymous SNV | / | 1/1 |
| 23 | chr3 | 75788109 | rs796849627 | ZNF717 | G | A | nonsynonymous SNV | / | 1/1 |
| 24 | chr3 | 75788130 | rs113708852 | ZNF717 | C | T | nonsynonymous SNV | / | 1/1 |
| 25 | chr3 | 75788137 | rs199883677 | ZNF717 | C | T | nonsynonymous SNV | / | 1/1 |
| 26 | chr3 | 184017075 | rs182086670 | PSMD2 | C | G | nonsynonymous SNV | 0/1 | 1/1 |
| 27 | chr4 | 80905990 | rs12647691 | ANTXR2 | C | G | nonsynonymous SNV | 1/1 | 0/1 |
| 28 | chr6 | 42075069 | rs55772414 | C6orf132 | A | G | nonsynonymous SNV | 0/1 | 1/1 |
| 29 | chr7 | 93540153 | rs17243826 | GNGT1 | G | C | nonsynonymous SNV | 1/1 | 0/1 |
| 30 | chr8 | 8750243 | MFHAS1 | A | G | nonsynonymous SNV | 0/0 | 0/1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukhtarova, K.; Zholdybayeva, E.; Kairov, U.; Akhmetollayev, I.; Nurimanov, C.; Kulmirzayev, M.; Makhambetov, Y.; Ramankulov, Y. Whole-Exome Sequencing Reveals Pathogenic SIRT1 Variant in Brain Arteriovenous Malformation: A Case Report. Genes 2022, 13, 1689. https://doi.org/10.3390/genes13101689
Mukhtarova K, Zholdybayeva E, Kairov U, Akhmetollayev I, Nurimanov C, Kulmirzayev M, Makhambetov Y, Ramankulov Y. Whole-Exome Sequencing Reveals Pathogenic SIRT1 Variant in Brain Arteriovenous Malformation: A Case Report. Genes. 2022; 13(10):1689. https://doi.org/10.3390/genes13101689
Chicago/Turabian StyleMukhtarova, Kymbat, Elena Zholdybayeva, Ulykbek Kairov, Ilyas Akhmetollayev, Chingiz Nurimanov, Marat Kulmirzayev, Yerbol Makhambetov, and Yerlan Ramankulov. 2022. "Whole-Exome Sequencing Reveals Pathogenic SIRT1 Variant in Brain Arteriovenous Malformation: A Case Report" Genes 13, no. 10: 1689. https://doi.org/10.3390/genes13101689
APA StyleMukhtarova, K., Zholdybayeva, E., Kairov, U., Akhmetollayev, I., Nurimanov, C., Kulmirzayev, M., Makhambetov, Y., & Ramankulov, Y. (2022). Whole-Exome Sequencing Reveals Pathogenic SIRT1 Variant in Brain Arteriovenous Malformation: A Case Report. Genes, 13(10), 1689. https://doi.org/10.3390/genes13101689






