Differentially Methylated DNA Regions and Left Ventricular Hypertrophy in African Americans: A HyperGEN Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Epigenome Analysis
2.3. Covariates
2.4. Statistical Analysis
2.5. Replication
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnett, D.K.; Hong, Y.; Bella, J.N.; Oberman, A.; Kitzman, D.W.; Hopkins, P.N.; Rao, D.; Devereux, R.B. Sibling correlation of left ventricular mass and geometry in hypertensive African Americans and whites: The HyperGEN study. Am. J. Hypertens. 2001, 14, 1226–1230. [Google Scholar] [CrossRef]
- Drazner, M.H.; Dries, D.L.; Peshock, R.M.; Cooper, R.S.; Klassen, C.; Kazi, F.; Willett, D.; Victor, R.G. Left ventricular hypertrophy is more prevalent in blacks than whites in the general population: The Dallas Heart Study. Hypertension 2005, 46, 124–129. [Google Scholar] [CrossRef]
- Kizer, J.R.; Arnett, D.K.; Bella, J.N.; Paranicas, M.; Rao, D.C.; Province, M.A.; Oberman, A.; Kitzman, D.W.; Hopkins, P.N.; Liu, J.E.; et al. Differences in left ventricular structure between black and white hypertensive adults: The Hypertension Genetic Epidemiology Network study. Hypertension 2004, 43, 1182–1188. [Google Scholar] [CrossRef]
- Skelton, T.N.; Andrew, M.E.; Arnett, D.K.; Burchfiel, C.M.; Garrison, R.J.; Samdarshi, T.E.; Taylor, H.A.; Hutchinson, R.G. Echocardiographic left ventricular mass in African-Americans: The Jackson cohort of the Atherosclerosis Risk in Communities Study. Echocardiography 2003, 20, 111–120. [Google Scholar] [CrossRef]
- Levy, D.; Garrison, R.J.; Savage, D.D.; Kannel, W.B.; Castelli, W.P. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 1990, 322, 1561–1566. [Google Scholar] [CrossRef]
- Schillaci, G.; Verdecchia, P.; Porcellati, C.; Cuccurullo, O.; Cosco, C.; Perticone, F. Continuous relation between left ventricular mass and cardiovascular risk in essential hypertension. Hypertension 2000, 35, 580–586. [Google Scholar] [CrossRef]
- Benjamin, E.J.; Levy, D. Why is left ventricular hypertrophy so predictive of morbidity and mortality? Am. J. Med. Sci. 1999, 317, 168–175. [Google Scholar] [CrossRef]
- Vakili, B.A.; Okin, P.M.; Devereux, R.B. Prognostic implications of left ventricular hypertrophy. Am. Heart J. 2001, 141, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Koren, M.J.; Devereux, R.B.; Casale, P.N.; Savage, D.D.; Laragh, J.H. Relation of left ventricular mass and geometry to morbidity and mortality in uncomplicated essential hypertension. Ann. Intern. Med. 1991, 114, 345–352. [Google Scholar] [CrossRef]
- Levy, D. Left ventricular hypertrophy and cardiovascular disease risk. In Hypertension Primer, 4th ed.; Izzo, J.L., Sica, D.A., Jr., Black, H.R., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008; pp. 254–257. [Google Scholar]
- Stewart, M.H.; Lavie, C.J.; Shah, S.; Englert, J.; Gilliland, Y.; Qamruddin, S.; Dinshaw, H.; Cash, M.; Ventura, H.; Milani, R. Prognostic Implications of Left Ventricular Hypertrophy. Prog. Cardiovasc. Dis. 2018, 61, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Berry, J.D.; Ayers, C.R.; Peshock, R.M.; Khera, A.; de Lemos, J.A.; Patel, P.C.; Markham, D.W.; Drazner, M.H. Left ventricular hypertrophy, aortic wall thickness, and lifetime predicted risk of cardiovascular disease: The Dallas Heart Study. JACC Cardiovasc. Imaging. 2010, 3, 605–613. [Google Scholar] [CrossRef]
- Bluemke, D.A.; Kronmal, R.A.; Lima, J.A.C.; Liu, K.; Olson, J.; Burke, G.L.; Folsom, A.R. The relationship of left ventricular mass and geometry to incident cardiovascular events: The MESA study. JACC 2008, 52, 2148–2155. [Google Scholar] [CrossRef]
- Gardin, J.M.; Wagenknecht, L.E.; Anton-Culver, H.; Flack, J.; Gidding, S.; Kurosaki, T.; Wong, N.D.; Manolio, T.A. Relationship of cardiovascular risk factors to echocardiographic left ventricular mass in healthy young black and white adult men and women. The CARDIA study. Circulation 1995, 92, 380–387. [Google Scholar] [CrossRef]
- De Simone, G.; Tang, W.; Devereux, R.B.; Hunt, S.C.; Kitzman, D.W.; Rao, D.; Arnett, D.K. Assessment of the interaction of heritability of volume load and left ventricular mass: The HyperGEN offspring study. J. Hypertens. 2007, 25, 1397–1402. [Google Scholar] [CrossRef]
- Bella, J.N.; MacCluer, J.W.; Roman, M.J.; Almasy, L.; North, K.E.; Best, L.G.; Lee, E.T.; Fabsitz, R.R.; Howard, B.V.; Devereux, R.B. Heritability of left ventricular dimensions and mass in American Indians: The Strong Heart Study. J. Hypertens. 2004, 22, 281–286. [Google Scholar] [CrossRef]
- Jin, Y.; Kuznetsova, T.; Bochud, M.; Richart, T.; Thijs, L.; Cusi, D.; Fagard, R.; Staessen, J.A. Heritability of left ventricular structure and function in Caucasian families. Eur. J. Echocardiogr. 2011, 12, 326–332. [Google Scholar]
- Alame, A.J.; Garg, S.; Kozlitina, J.; Ayers, C.; Peshock, R.M.; Matulevicius, S.A.; Drazner, M.H. Association of African ancestry with electrocardiographic voltage and concentric left ventricular hypertrophy: The Dallas Heart Study. JAMA Cardiolo. 2018, 3, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Arnett, D.K.; Devereux, R.B.; Atwood, L.D.; Kitzman, D.W.; Rao, D.C. Linkage of left ventricular early diastolic peak filling velocity to chromosome 5 in hypertensive African Americans: The HyperGEN echocardiography study. Am. J. Hypertens. 2002, 15, 621–627. [Google Scholar] [CrossRef]
- Arnett, D.K.; Devereux, R.B.; Rao, D.C.; Li, N.; Tang, W.; Kraemer, R.; Claas, S.A.; Leon, J.M.; Broeckel, U. Novel genetic variants contributing to left ventricular hypertrophy: The HyperGEN study. J. Hypertens. 2009, 27, 1585–1593. [Google Scholar] [CrossRef]
- Fox, E.R.; Klos, K.L.; Penman, A.D.; Blair, G.J.; Blossom, B.D.; Arnett, D.; Devereux, R.B.; Samdarshi, T.; Boerwinkle, E.; Mosley, T.H., Jr. Heritability and genetic linkage of left ventricular mass, systolic and diastolic function in hypertensive African Americans (From the GENOA Study). Am. J. Hypertens. 2010, 23, 870–875. [Google Scholar] [CrossRef]
- Della-Morte, D.; Beecham, A.; Rundek, T.; Wang, L.; McClendon, M.S.; Slifer, S.; Blanton, S.H.; Di Tullio, M.R.; Sacco, R.L. A follow-up study for left ventricular mass on chromosome 12p11 identifies potential candidate genes. BMC Med. Genet. 2011, 12, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, A.I.; Tang, W.; Shi, G.; Devereux, R.B.; Eckfeldt, J.H.; Arnett, D.K. Epistatic effects of ACE I/D and AGT gene variants on left ventricular mass in hypertensive patients: The HyperGEN study. J. Hum. Hypertens. 2012, 26, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Arnett, D.K.; Meyers, K.J.; Devereux, R.B.; Tiwari, H.K.; Gu, C.C.; Vaughan, L.K.; Perry, R.T.; Patki, A.; Claas, S.A.; Sun, Y.V.; et al. Genetic variation in NCAM1 contributes to left ventricular wall thickness in hypertensive families. Circ. Res. 2011, 108, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Arnett, D.K.; Li, N.; Tang, W.; Rao, D.C.; Devereux, R.B.; Claas, S.A.; Kraemer, R.; Broeckel, U. Genome-wide association study identifies single-nucleotide polymorphism in KCNB1 associated with left ventricular mass in humans: The HyperGEN Study. BMC Med. Genet. 2009, 10, 43. [Google Scholar] [CrossRef]
- Vasan, R.S.; Larson, M.G.; Aragam, J.; Wang, T.J.; Mitchell, G.F.; Kathiresan, S.; Newton-Cheh, C.; Vita, J.A.; Keyes, M.J.; O’Donnell, C.J.; et al. Genome-wide association of echocardiographic dimensions, brachial artery endothelial function and treadmill exercise responses in the Framingham Heart Study. BMC Med. Genet. 2007, 8 (Suppl. S1), S2. [Google Scholar] [CrossRef]
- Vasan, R.S.; Glazer, N.L.; Felix, J.F.; Lieb, W.; Wild, P.S.; Felix, S.B.; Watzinger, N.; Larson, M.G.; Smith, N.L.; Dahghan, A.; et al. Genetic variants associated with cardiac structure and function: A meta-analysis and replication of genome-wide association data. JAMA 2009, 302, 168–178. [Google Scholar] [CrossRef]
- Dekkers, C.; Treiber, F.A.; Kapuku, G.; van den Oord, E.J.C.G.; Sneider, H. Growth of left ventricular mass in African American and European American youth. Hypertension 2002, 39, 943–951. [Google Scholar] [CrossRef]
- Zhang, T.; Li, S.; Bazzano, L.; He, J.; Whelton, P.; Chen, W. Trajectories of childhood blood pressure and adult left ventricular hypertrophy: The Bogalusa Heart Study. Hypertension 2018, 72, 93–101. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Tingare, A.; Thienpont, B.; Roderick, H.L. Epigenetics in the heart: The role of histone modifications in cardiac remodelling. Biochem. Soc. Trans. 2013, 41, 789–796. [Google Scholar] [CrossRef]
- Xiao, D.; Dasgupta, C.; Chen, M.; Zhang, K.; Buchholz, J.; Xu, Z.; Zhang, L. Inhibition of DNA methylation reverses norepinephrine-induced cardiac hypertrophy in rats. Cardiovasc. Res. 2014, 101, 373–382. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.; Keshvani, N.; Ayers, C.; Correa, A.; Drazner, M.H.; Lewis, A.; Rodriguez, C.J.; Hall, M.E.; Fox, E.R.; Mentz, R.J.; et al. Association of cardiac injury and malignant left ventricular hypertrophy with risk of heart failure in African Americans: The Jackson Heart Study. JAMA Cardiol. 2019, 4, 51–58. [Google Scholar] [CrossRef]
- Heidenreich, P.A.; Albert, N.M.; Allen, L.A.; Bluemke, D.A.; Butler, J.; Fonarow, G.C.; Ikonomidis, J.S.; Khavjou, O.; Konstam, M.A.; Maddox, T.M.; et al. Forecasting the impact of heart failure in the United States: A policy statement from the American Heart Association. Circ. Heart Fail. 2013, 6, 606–619. [Google Scholar] [CrossRef]
- William, R.R.; Rao, D.C.; Ellison, R.C.; Arnett, D.K.; Heiss, G.; Oberman, A.; Eckfeldt, J.H.; Leppert, M.F.; Province, M.A.; Mockrin, S.C.; et al. NHLBI family blood pressure program: Methodology and recruitment in the HyperGEN network. Ann. Epidemiol. 2000, 10, 389–400. [Google Scholar] [CrossRef]
- Devereux, R.B.; Roman, M.J. Evaluation of cardiac function and vascular structure and function by echocardiography and other noninvasive techniques. In Hypertension: Pathophysiology, Diagnosis, and Management; Laragh, J.H., Brenne, B.M., Eds.; Raven P: New York, NY, USA, 1995; pp. 1969–1985. [Google Scholar]
- Akinyemiju, T.; Do, A.N.; Patki, A.; Aslibekyan, S.; Zhi, D.; Hidalgo, B.; Tiwari, H.K.; Absher, D.; Geng, X.; Arnett, D.K.; et al. Epigenome-wide association study of metabolic syndrome in African-American adults. Clin. Epigenet. 2018, 10, 49. [Google Scholar] [CrossRef]
- Bjørnland, T.; Bye, A.; Ryeng, E.; Wisløff, U.; Langaas, M. Powerful extreme phenotype sampling designs and score tests for genetic association studies. Stat. Med. 2018, 37, 4234–4251. [Google Scholar] [CrossRef]
- Zhou, W.; Triche Jr, T.J.; Laird, P.W.; Shen, H. SeSAMe: Reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018, 46, e123. [Google Scholar] [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef]
- Price, A.L.; Weale, M.E.; Patterson, N.; Myers, S.R.; Need, A.C.; Shianna, K.V.; Ge, D.; Rotter, J.I.; Torres, E.; Taylor, K.D.; et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008, 83, 132–135. [Google Scholar] [CrossRef]
- Gomez, L.; Odom, G.J.; Young, J.I.; Martin, E.R.; Liu, L.; Chen, X.; Griswold, A.J.; Gao, Z.; Zhang, L.; Wang, L. coMethDMR: Accurate identification of co-methylated and differentially methylated regions in epigenome-wide association studies with continuous phenotypes. Nucleic Acids Res. 2019, 47, e98. [Google Scholar] [CrossRef]
- Taylor, H.A., Jr.; Wilson, J.G.; Jones, D.W.; Sarpong, D.F.; Srinivasan, A.; Garrison, R.J.; Nelson, C.; Wyatt, S.B. Toward resolution of cardiovascular health disparities in African Americans: Design and methods of the Jackson Heart Study. Ethn. Dis. 2005, 15, S6-4. [Google Scholar]
- Raffield, L.M.; Lu, A.T.; Szeto, M.D.; Little, A.; Grinde, K.E.; Shaw, J.; Auer, P.L.; Cushman, M.; Horvath, S.; Irvin, M.R.; et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, TOPMed Hematology & Hemostasis Working Group, Zakai NA, Reiner AP. Coagulation factor VIII: Relationship to cardiovascular disease risk and whole genome sequence and epigenome-wide analysis in African Americans. J. Thromb. Haemost. 2020, 18, 1335–1347. [Google Scholar]
- Daniels, P.R.; Kardia, S.L.; Hanis, C.L.; Brown, C.A.; Hutchinson, R.; Boerwinkle, E.; Turner, S.T. Genetic Epidemiology Network of Arteriopathy study. Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. Am. J. Med. 2004, 116, 676–681. [Google Scholar] [CrossRef]
- Parry, H.M.; Donnelly, L.A.; Van Zuydam, N.; Doney, A.S.; Elder, D.H.; Morris, A.D.; Struthers, A.D.; Palmer, C.N.; Lang, C.C. Wellcome Trust Case Control Consortium 2. Genetic variants predicting left ventricular hypertrophy in a diabetic population: A Go-DARTS study including meta-analysis. Cardiovasc. Diabetol. 2013, 12, 109. [Google Scholar] [CrossRef]
- Sano, M.; Kamitsuji, S.; Kamatani, N.; Tabara, Y.; Kawaguchi, T.; Matsuda, F.; Yamagishi, H.; Fukuda, K. JPDSC Genome-wide association study of absolute QRS voltage identifies common variants of TBX3 as genetic determinants of left ventricular mass in a healthy Japanese population. PLoS ONE 2016, 11, e0155550. [Google Scholar] [CrossRef]
- Hong, K.W.; Shin, D.J.; Lee, S.H.; Son, N.H.; Go, M.J.; Lim, J.E.; Shin, C.; Jang, Y.; Oh, B. Common variants in RYR1 are associated with left ventricular hypertrophy assessed by electrocardiogram. Eur. Heart J. 2012, 33, 1250–1256. [Google Scholar] [CrossRef]
- Ung, C.; Sanchez, A.V.; Shen, L.; Davoudi, S.; Ahmadi, T.; Navarro-Gomez, D.; Chen, C.J.; Hancock, H.; Penman, A.; Hoadley, S.; et al. Whole exome sequencing identification of novel candidate genes in patients with proliferative diabetic retinopathy. Vision Res. 2017, 139, 168–176. [Google Scholar] [CrossRef]
- Meier, N.; Bruder, E.; Lapaire, O.; Hoesli, I.; Kang, A.; Hench, J.; Hoeller, S.; De Geyter, J.; Miny, P.; Heinimann, K.; et al. Exome sequencing of fetal anomaly syndromes: Novel phenotype-genotype discoveries. Eur. J. Hum. Genet. 2019, 27, 730–737. [Google Scholar] [CrossRef]
- Wu, S.; Hivert, M.F.; Cardenas, A.; Zhong, J.; Rifas-Shiman, S.L.; Agha, G.; Colicino, E.; Just, A.C.; Amarasiriwardena, C.; Lin, X.; et al. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in Project Viva: An epigenome-wide association study. Environ. Health Perspect. 2017, 125, 087019. [Google Scholar] [CrossRef]
- Zheng, M.L.; Du, X.P.; Zhao, L.; Yang, X.C. Expression profile of circular RNAs in epicardial adipose tissue in heart failure. Chin. Med. J. 2020, 133, 2565–2572. [Google Scholar] [CrossRef]
- Arpón, A.; Milagro, F.I.; Ramos-Lopez, O.; Mansego, M.L.; Santos, J.L.; Riezu-Boj, J.I.; Martínez, J.A. Epigenome-wide association study in peripheral white blood cells involving insulin resistance. Sci. Rep. 2019, 9, 2445. [Google Scholar] [CrossRef]
- Mancusi, C.; Izzo, R.; di Gioia, G.; Losi, M.A.; Barbato, E.; Morisco, C. Insulin Resistance the Hinge Between Hypertension and Type 2 Diabetes. High Blood Press Cardiovasc. Prev. 2020, 27, 515–526. [Google Scholar] [CrossRef]
- Kianu Phanzu, B.; Nkodila Natuhoyila, A.; Kintoki Vita, E.; M’Buyamba Kabangu, J.R.; Longo-Mbenza, B. Association between insulin resistance and left ventricular hypertrophy in asymptomatic, Black, sub-Saharan African, hypertensive patients: A case-control study. BMC Cardiovasc. Disord. 2021, 21, 1. [Google Scholar] [CrossRef]
- Sciacqua, A.; Cimellaro, A.; Mancuso, L.; Miceli, S.; Cassano, V.; Perticone, M.; Fiorentino, T.V.; Andreozzi, F.; Succurro, E.; Sesti, G.; et al. Different Patterns of Left Ventricular Hypertrophy in Metabolically Healthy and Insulin-Resistant Obese Subjects. Nutrients 2020, 12, 412. [Google Scholar] [CrossRef]
- Al-Daydamony, M.M.; El-Tahlawi, M. What Is the Effect of Metabolic Syndrome without Hypertension on Left Ventricular Hypertrophy? Echocardiography 2016, 33, 1284–1289. [Google Scholar] [CrossRef]
- Pang, B.; Hu, C.; Wu, G.; Zhang, Y.; Lin, G. Identification of Target Genes in Hypertension and Left Ventricular Remodeling. Medicine 2020, 99, e21195. [Google Scholar] [CrossRef]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef] [Green Version]


| N (%)/Mean (SD) | HyperGEN (N = 611) | JHS (N = 1054) | GENOA (N = 839) |
|---|---|---|---|
| Age, years | 48.4 (11.1) | 53.7 (12.1) | 62.7 (9.6) |
| Male | 205 (33.4%) | 378 (35.9%) | 242 (28.8%) |
| Body mass index, kg/m2 | 32.4 (8.2) | 32.0 (7.2) | 31.4 (6.6) |
| Systolic blood pressure, mmHg | 131.6 (23.6) | 126.7 (16.1) | 143.2 (23.4) |
| Diastolic blood pressure, mmHg | 75.5 (12.6) | 76.2 (8.5) | 79.3 (11.5) |
| Hypertension, % | 459 (75.1%) | 205 (19.4%) | 660 (78.7%) |
| LV mass, g (LVM) | 180.0 (63.3) | 151.6 (41.1) | 160.1 (48.6) |
| LV mass indexed to height, g/m (LVMHT27) | 45.0 (16.1) | 36.9 (9.8) | 39.3 (11.3) |
| Relative wall thickness, cm (RWT) | 0.3 (0.1) | 0.4 (0.1) | 0.3 (0.1) |
| LV internal diastolic dimension, cm (LVIDD) | 5.2 (0.6) | - | 5.2 (0.5) |
| Left atrial systolic dimension, cm (LASD) | 3.4 (0.6) | - | 3.6 (0.5) |
| Ejection fraction, % (EF) | 61.8 (6.9) | 62.1 (7.8) | 60.6 (7.7) |
| Midwall shortening, % (MWS) | 17.0 (2.7) | - | 17.6 (2.4) |
| Trait | Chr | nCpGs | DMR (bp) | Location | Gene | HyperGEN | JHS | ||
|---|---|---|---|---|---|---|---|---|---|
| β(SE) | FDR | β(SE) | FDR | ||||||
| EF | 8 | 3 | 59 | Open Sea | XKR6 | 0.0048(0.0012) | 0.056 | 0.0010(0.0013) | 1 |
| LVIDD | 3 | 5 | 335 | Open Sea | TRAK1 | 0.1130(0.0270) | 0.042 | -- | -- |
| LVMHT27 | 16 | 3 | 247 | Island | GSE1 | −0.0021(0.0005) | 0.085 | 0.0003(0.0005) | 0.61 |
| 16 | 5 | 225 | Island | RPS15 A | −0.0018(0.0005) | 0.085 | −0.0001(0.0005) | 0.75 | |
| 16 | 4 | 350 | Island | PSMD7 | −0.0026(0.0007) | 0.085 | −0.0012(0.0012) | 1 | |
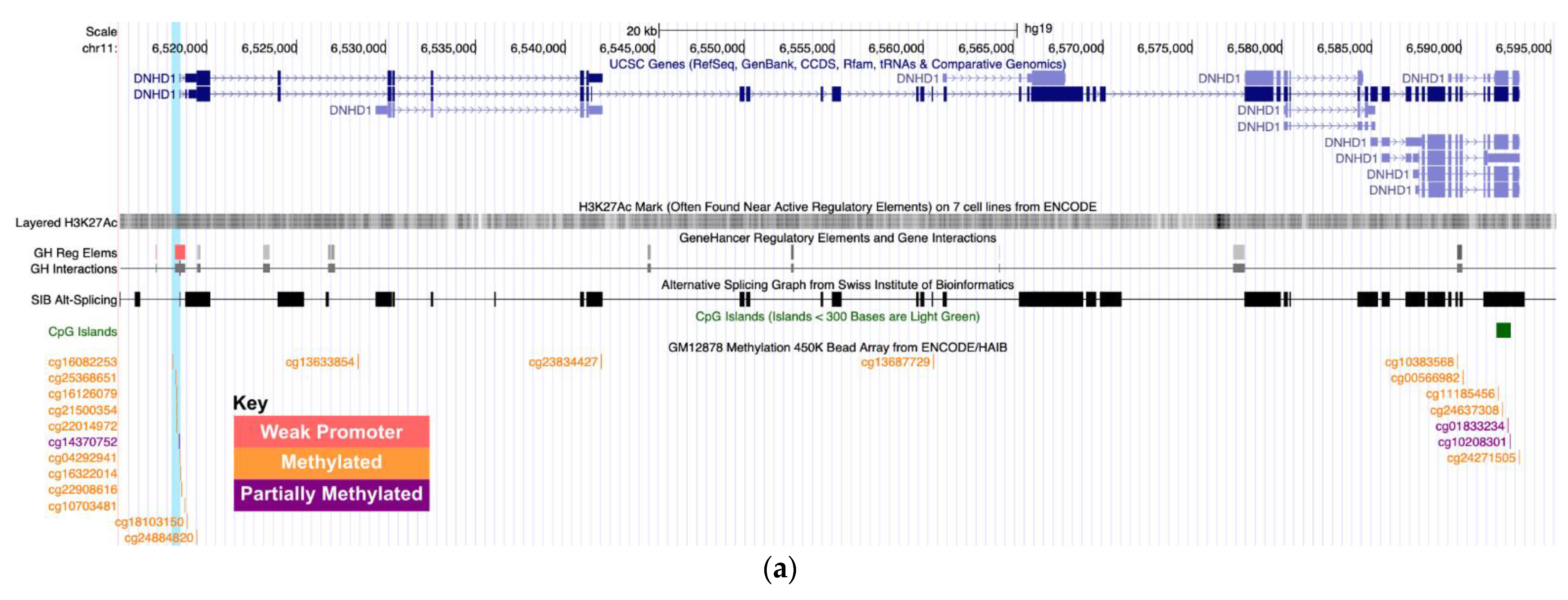
| RWT | 11 | 9 | 469 | Open Sea | DNHD1 | −1.2688(0.2749) | 0.008 | −0.1598(0.1781) | 0.37 |
| Trait | Chr | Gene | CpG | HyperGEN | JHS | GENOA | |||
|---|---|---|---|---|---|---|---|---|---|
| β | FDR | β | FDR | β | FDR | ||||
| EF | 8 | XKR6 | cg27534424 | 0.0002 | 1 | 1.18 × 10−4 | 0.17 | 1.24 × 10−5 | 0.89 |
| cg09051192 | 0.0001 | 1 | 7.01 × 10−5 | 0.23 | 3.69 × 10−5 | 0.80 | |||
| cg23673360 | 9.76 × 10−5 | 1 | 7.26 × 10−5 | 0.19 | −5.62 × 10−6 | 0.89 | |||
| LVIDD | 3 | TRAK1 | cg01855070 | 0.013 | 0.83 | -- | -- | −0.0038 | 0.89 |
| cg08804892 | 0.011 | 1 | -- | -- | −0.0008 | 0.89 | |||
| cg08508763 | 0.015 | 0.93 | -- | -- | −0.0037 | 0.89 | |||
| cg23715029 | 0.023 | 0.06 | -- | -- | −0.0017 | 0.89 | |||
| cg03168947 | 0.019 | 0.32 | -- | -- | −0.0043 | 0.89 | |||
| LVMHT27 | 16 | GSE1 | cg02075099 | −4.60 × 10−5 | 1 | 6.43 × 10−6 | 0.58 | 1.39 × 10−6 | 0.89 |
| cg12551166 | −7.62 × 10−5 | 1 | 3.10 × 10−5 | 0.20 | 1.39 × 10−5 | 0.80 | |||
| cg14842398 | −4.36 × 10−5 | 1 | −1.49 × 10−5 | 0.58 | −1.24 × 10−5 | 0.89 | |||
| RPS15 A | cg07623022 | −1.51 × 10−5 | 1 | 7.73 × 10−6 | 0.58 | −1.92 × 10−5 | 0.80 | ||
| cg01527529 | −6.52 × 10−5 | 1 | −2.72 × 10−5 | 0.41 | −4.77 × 10−6 | 0.89 | |||
| cg21024145 | −4.42 × 10−5 | 1 | 3.71 × 10−5 | 0.32 | 4.50 × 10−6 | 0.89 | |||
| cg26593997 | −7.54 × 10−5 | 1 | 6.52 × 10−7 | 1 | 4.92 × 10−6 | 0.89 | |||
| cg01680999 | −4.76 × 10−5 | 1 | −1.72 × 10−5 | 0.56 | −9.40 × 10−6 | 0.89 | |||
| PSMD7 | cg00504756 | −7.08 × 10−5 | 1 | 1.99 × 10−5 | 0.52 | 4.82 × 10−5 | 0.80 | ||
| cg07773642 | −4.86 × 10−5 | 1 | 2.36 × 10−5 | 0.72 | 2.08 × 10−6 | 0.89 | |||
| cg13135456 | −5.94 × 10−5 | 1 | −2.26 × 10−5 | 0.37 | −3.99 × 10−5 | 0.80 | |||
| cg08598838 | −3.14 × 10−5 | 1 | −3.32 × 10−5 | 0.19 | −3.67 × 10−6 | 0.89 | |||
| RWT | 11 | DNHD1 | cg14370752 | −0.156 | 1 | -- | -- | −0.0138 | 0.89 |
| cg16322014 | −0.131 | 0.25 | −0.016 | 0.37 | −0.0020 | 0.89 | |||
| cg22908616 | −0.153 | 0.08 | −0.052 | 0.17 | −0.0309 | 0.80 | |||
| cg16082253 | −0.079 | 1 | 0.021 | 0.32 | 0.0040 | 0.89 | |||
| cg25368651 | −0.110 | 0.10 | −0.031 | 0.19 | −0.0102 | 0.80 | |||
| cg16126079 | −0.121 | 0.08 | −0.026 | 0.21 | −0.0044 | 0.89 | |||
| cg21500354 | −0.139 | 0.11 | −0.027 | 0.33 | −0.0141 | 0.80 | |||
| cg22014972 | −0.127 | 0.19 | −0.035 | 0.19 | −0.0116 | 0.80 | |||
| cg04292941 | −0.083 | 1 | −0.021 | 0.52 | −0.0144 | 0.89 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, A.C.; Patki, A.; Claas, S.A.; Tiwari, H.K.; Chaudhary, N.S.; Absher, D.M.; Lange, L.A.; Lange, E.M.; Zhao, W.; Ratliff, S.M.; et al. Differentially Methylated DNA Regions and Left Ventricular Hypertrophy in African Americans: A HyperGEN Study. Genes 2022, 13, 1700. https://doi.org/10.3390/genes13101700
Jones AC, Patki A, Claas SA, Tiwari HK, Chaudhary NS, Absher DM, Lange LA, Lange EM, Zhao W, Ratliff SM, et al. Differentially Methylated DNA Regions and Left Ventricular Hypertrophy in African Americans: A HyperGEN Study. Genes. 2022; 13(10):1700. https://doi.org/10.3390/genes13101700
Chicago/Turabian StyleJones, Alana C., Amit Patki, Steven A. Claas, Hemant K. Tiwari, Ninad S. Chaudhary, Devin M. Absher, Leslie A. Lange, Ethan M. Lange, Wei Zhao, Scott M. Ratliff, and et al. 2022. "Differentially Methylated DNA Regions and Left Ventricular Hypertrophy in African Americans: A HyperGEN Study" Genes 13, no. 10: 1700. https://doi.org/10.3390/genes13101700
APA StyleJones, A. C., Patki, A., Claas, S. A., Tiwari, H. K., Chaudhary, N. S., Absher, D. M., Lange, L. A., Lange, E. M., Zhao, W., Ratliff, S. M., Kardia, S. L. R., Smith, J. A., Irvin, M. R., & Arnett, D. K. (2022). Differentially Methylated DNA Regions and Left Ventricular Hypertrophy in African Americans: A HyperGEN Study. Genes, 13(10), 1700. https://doi.org/10.3390/genes13101700







