A Novel Single-Nucleotide Polymorphism in WNT4 Promoter Affects Its Transcription and Response to FSH in Chicken Follicles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collections
2.2. Cell Culture
2.3. Construction of WNT4 Promoter Deletion Vectors and Site-Directed Mutation
2.4. Cell Transfection and Luciferase Assay
2.5. SNP Identification, Polymorphism and Association Analysis
2.6. Electrophoretic Mobility Shift Assay (EMSA)
2.7. Real-Time Quantitative PCR
2.8. Statistical Analyses
3. Results
3.1. Critical Region of WNT4 Gene Response to FSH in Chicken GCs
3.2. Polymorphisms in the Critical Promoter Region of the Chicken WNT4 Gene
3.3. The Association of SNP−3015 (G > C) of WNT4 Gene with Chicken Production Traits
3.4. The Effects of SNP−3015 (G > C) on the Promoter Activity of the Chicken WNT4 Gene
3.5. SNP−3015 (G > C) Affects NFAT5 Binding in Chicken WNT4 Promoter
3.6. The Expression Level of WNT4 mRNA in Different Genotype Chicken Individuals
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cadigan, K.M.; Nusse, R. Wnt signaling: A common theme in animal development. Genes Dev. 1997, 11, 3286–3305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Waghmare, I.; Page-McCaw, A. Wnt Signaling in Stem Cell Maintenance and Differentiation in the Drosophila Germarium. Genes 2018, 9, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Amerongen, R.; Nusse, R. Towards an integrated view of Wnt signaling in development. Development 2009, 136, 3205–3214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell. Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef]
- van Amerongen, R.; Mikels, A.; Nusse, R. Alternative wnt signaling is initiated by distinct receptors. Sci. Signal 2008, 1, re9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz-Romond, T. Three decades of Wnt signalling. EMBO J. 2012, 31, 2664. [Google Scholar] [CrossRef] [Green Version]
- Boyer, A.; Lapointe, E.; Zheng, X.; Cowan, R.G.; Li, H.; Quirk, S.M.; DeMayo, F.J.; Richards, J.S.; Boerboom, D. WNT4 is required for normal ovarian follicle development and female fertility. FASEB J. 2010, 24, 3010–3025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, J.S. From Follicular Development and Ovulation to Ovarian Cancers: An Unexpected Journey. Vitam. Horm. 2018, 107, 453–472. [Google Scholar] [CrossRef]
- Dougherty, D.C.; Sanders, M.M. Estrogen action: Revitalization of the chick oviduct model. Trends Endocrinol. Metab. 2005, 16, 414–419. [Google Scholar] [CrossRef]
- Lim, C.H.; Lim, W.; Jeong, W.; Lee, J.Y.; Bae, S.M.; Kim, J.; Han, J.Y.; Bazer, F.W.; Song, G. Avian WNT4 in the female reproductive tracts: Potential role of oviduct development and ovarian carcinogenesis. PLoS ONE 2013, 8, e65935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chen, Q.; Liu, Z.; Guo, X.; Du, Y.; Yuan, Z.; Guo, M.; Kang, L.; Sun, Y.; Jiang, Y. Transcriptome Analysis on Single Small Yellow Follicles Reveals That Wnt4 Is Involved in Chicken Follicle Selection. Front. Endocrinol. 2017, 8, 317. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015, 94, 781–785. [Google Scholar] [CrossRef]
- Hsieh, M.; Johnson, M.A.; Greenberg, N.M.; Richards, J.S. Regulated expression of Wnts and Frizzleds at specific stages of follicular development in the rodent ovary. Endocrinology 2002, 143, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Ricken, A.; Lochhead, P.; Kontogiannea, M.; Farookhi, R. Wnt signaling in the ovary: Identification and compartmentalized expression of wnt-2, wnt-2b, and frizzled-4 mRNAs. Endocrinology 2002, 143, 2741–2749. [Google Scholar] [CrossRef] [PubMed]
- Harwood, B.N.; Cross, S.K.; Radford, E.E.; Haac, B.E.; De Vries, W.N. Members of the WNT signaling pathways are widely expressed in mouse ovaries, oocytes, and cleavage stage embryos. Dev. Dyn. 2008, 237, 1099–1111. [Google Scholar] [CrossRef]
- Wang, H.X.; Tekpetey, F.R.; Kidder, G.M. Identification of WNT/beta-CATENIN signaling pathway components in human cumulus cells. Mol. Hum. Reprod. 2009, 15, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Castañon, B.I.; Stapp, A.D.; Gifford, C.A.; Spicer, L.J.; Hallford, D.M.; Hernandez Gifford, J.A. Follicle-stimulating hormone regulation of estradiol production: Possible involvement of WNT2 and β-catenin in bovine granulosa cells. J. Anim. Sci. 2012, 90, 3789–3797. [Google Scholar] [CrossRef]
- Gupta, P.S.; Folger, J.K.; Rajput, S.K.; Lv, L.; Yao, J.; Ireland, J.J.; Smith, G.W. Regulation and regulatory role of WNT signaling in potentiating FSH action during bovine dominant follicle selection. PLoS ONE 2014, 9, e100201. [Google Scholar] [CrossRef] [Green Version]
- Franco, H.L.; Dai, D.; Lee, K.Y.; Rubel, C.A.; Roop, D.; Boerboom, D.; Jeong, J.W.; Lydon, J.P.; Bagchi, I.C.; Bagchi, M.K.; et al. WNT4 is a key regulator of normal postnatal uterine development and progesterone signaling during embryo implantation and decidualization in the mouse. FASEB J. 2011, 25, 1176–1187. [Google Scholar] [CrossRef] [Green Version]
- Angioni, S.; D’Alterio, M.N.; Coiana, A.; Anni, F.; Gessa, S.; Deiana, D. Genetic Characterization of Endometriosis Patients: Review of the Literature and a Prospective Cohort Study on a Mediterranean Population. Int. J. Mol. Sci. 2020, 21, 1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, S.S.; Hu, W.X.; Yang, T.L.; Chen, X.F.; Yan, H.; Chen, X.D.; Tan, L.J.; Tian, Q.; Deng, H.W.; Guo, Y. SNP-SNP interactions between WNT4 and WNT5A were associated with obesity related traits in Han Chinese Population. Sci. Rep. 2017, 7, 43939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Y.; Liu, Y.; Zhang, M.; Shan, Y.; Ji, G.; Ju, X.; Zou, J.; Shu, J. Identifying Signatures of Selection Related to Comb Development. J. Poult. Sci. 2021, 58, 5–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folsch, D.W.; Sulzer, B.; Meier, T.; Huber, H.U. The effect of the husbandry system on comb size and comb color in hens. Tierarztl. Prax. 1994, 22, 47–54. [Google Scholar]
- O’Connor, E.A.; Saunders, J.E.; Grist, H.; McLeman, M.A.; Wathes, C.M.; Abeyesinghe, S.M. The relationship between the comb and social behaviour in laying hens. Appl. Anim. Behav. Sci. 2011, 135, 293–299. [Google Scholar] [CrossRef]
- Aramburu, J.; Drews-Elger, K.; Estrada-Gelonch, A.; Minguillón, J.; Morancho, B.; Santiago, V.; López-Rodríguez, C. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem. Pharmacol. 2006, 72, 1597–1604. [Google Scholar] [CrossRef]
- O’Connor, R.S.; Mills, S.T.; Jones, K.A.; Ho, S.N.; Pavlath, G.K. A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 2007, 120, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Berga-Bolanos, R.; Drews-Elger, K.; Aramburu, J.; Lopez-Rodriguez, C. NFAT5 regulates T lymphocyte homeostasis and CD24-dependent T cell expansion under pathologic hypernatremia. J. Immunol. 2010, 185, 6624–6635. [Google Scholar] [CrossRef] [Green Version]
- Go, W.Y.; Liu, X.; Roti, M.A.; Liu, F.; Ho, S.N. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. USA 2004, 101, 10673–10678. [Google Scholar] [CrossRef] [Green Version]
- López-Rodríguez, C.; Antos, C.L.; Shelton, J.M.; Richardson, J.A.; Lin, F.; Novobrantseva, T.I.; Bronson, R.T.; Igarashi, P.; Rao, A.; Olson, E.N. Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc. Natl. Acad. Sci. USA 2004, 101, 2392–2397. [Google Scholar] [CrossRef] [Green Version]
- Mak, M.C.; Lam, K.M.; Chan, P.K.; Lau, Y.B.; Tang, W.H.; Yeung, P.K.K.; Ko, B.C.B.; Chung, S.M.S.; Chung, S.K. Embryonic lethality in mice lacking the nuclear factor of activated T cells 5 protein due to impaired cardiac development and function. PLoS ONE 2011, 6, e19186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saneyoshi, T.; Kume, S.; Amasaki, Y.; Mikoshiba, K. The Wnt calcium pathway activates NFAT and promotes ventral cell fate in Xenopus. Nature 2002, 417, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, F.; Yamamoto, H. Effect of inhibition of glycogen synthase kinase-3 on cardiac hypertrophy during acute pressure overload. Gen. Thorac. Cardiovasc. Surg. 2010, 58, 263–264. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zhou, Y.; Rychahou, P.; Liu, C.; Weiss, H.L.; Evers, B.M. NFAT5 represses canonical Wnt signaling via inhibition of beta-catenin acetylation and participates in regulating intestinal cell differentiation. Cell Death Dis. 2013, 4, e671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, H.; Xiong, Q.; Ji, Z.; Zhang, F.; Liu, Y.; Chen, M. NFAT5 is Regulated by p53/miR-27a Signal Axis and Promotes Mouse Ovarian Granulosa Cells Proliferation. Int. J. Biol. Sci. 2019, 15, 287–297. [Google Scholar] [CrossRef]
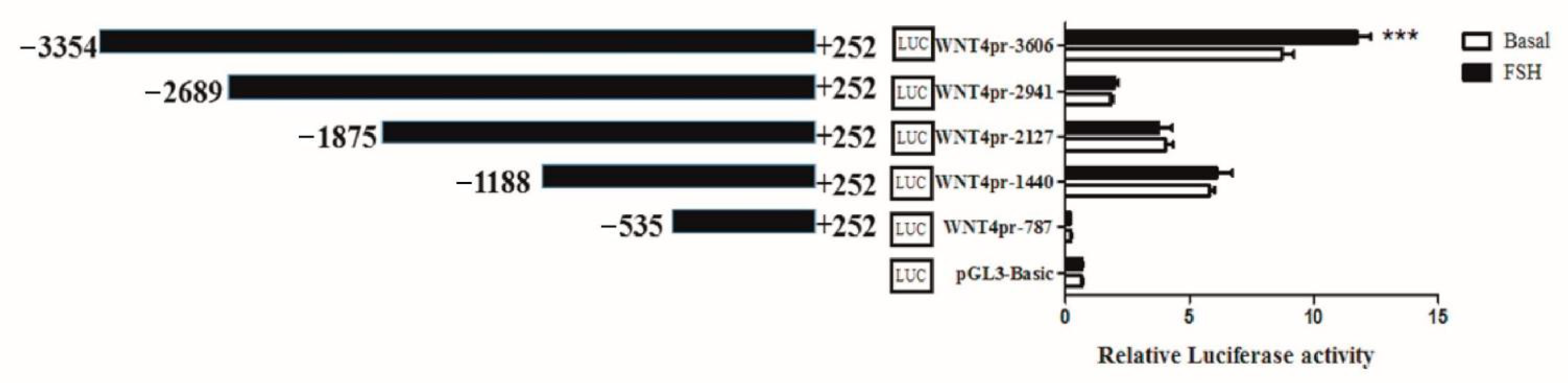
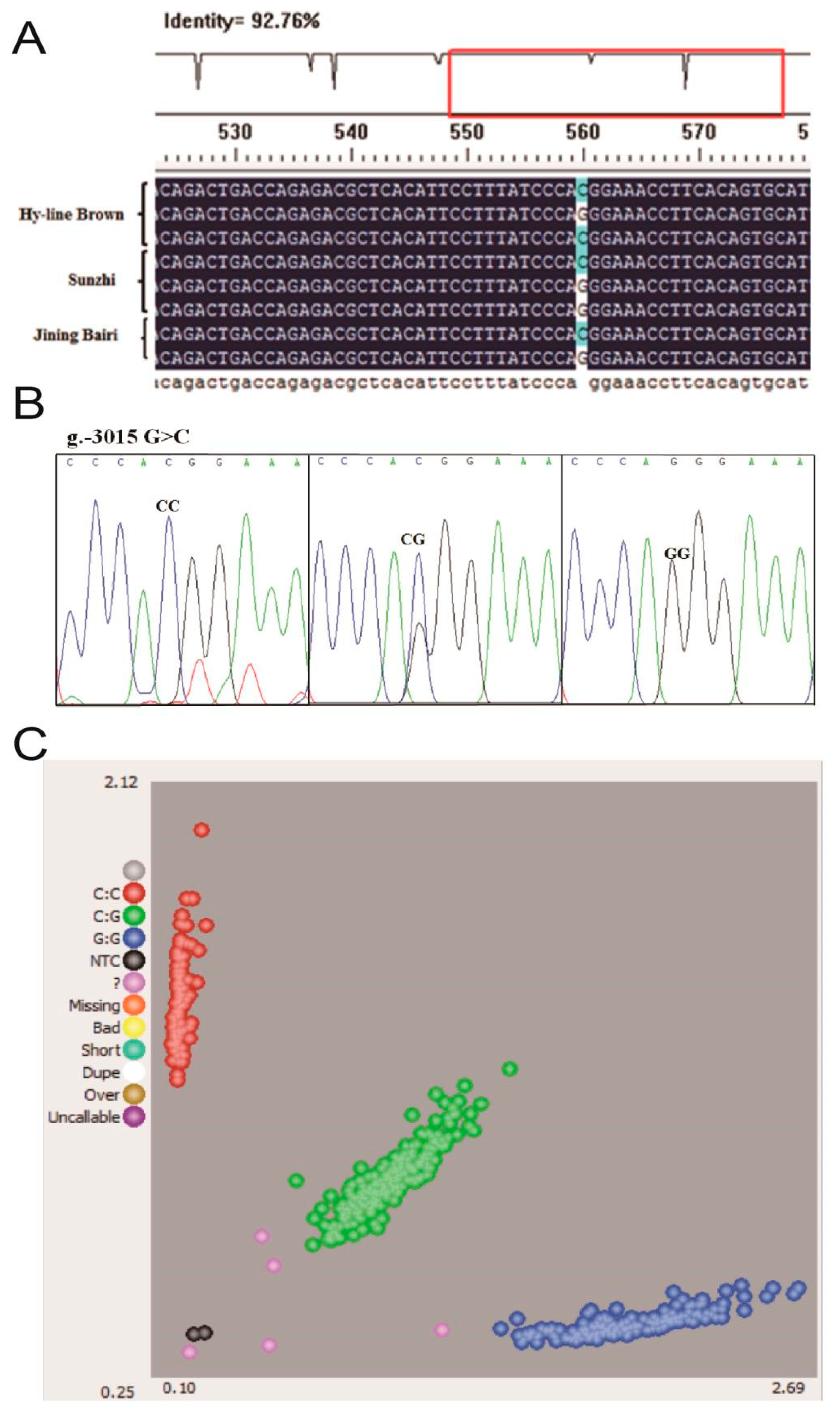

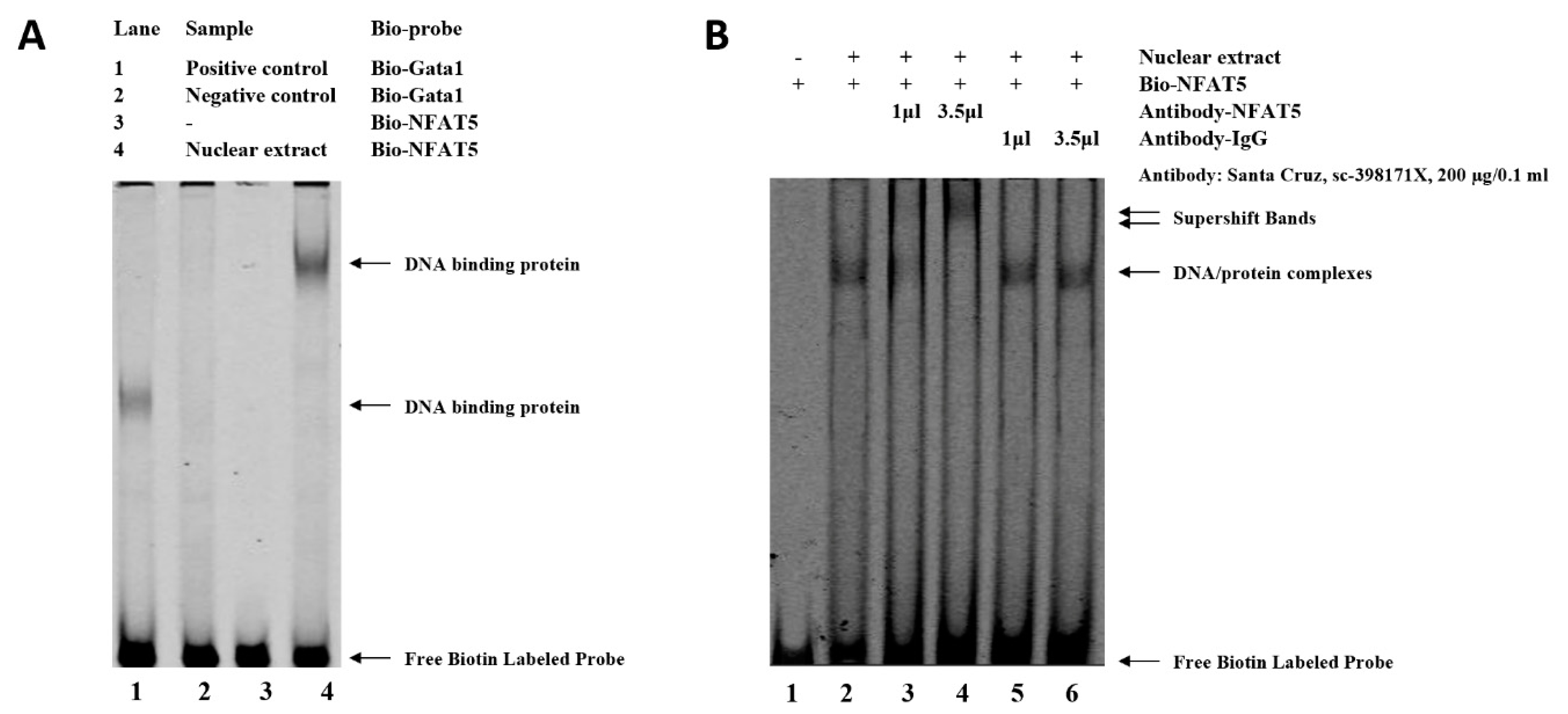
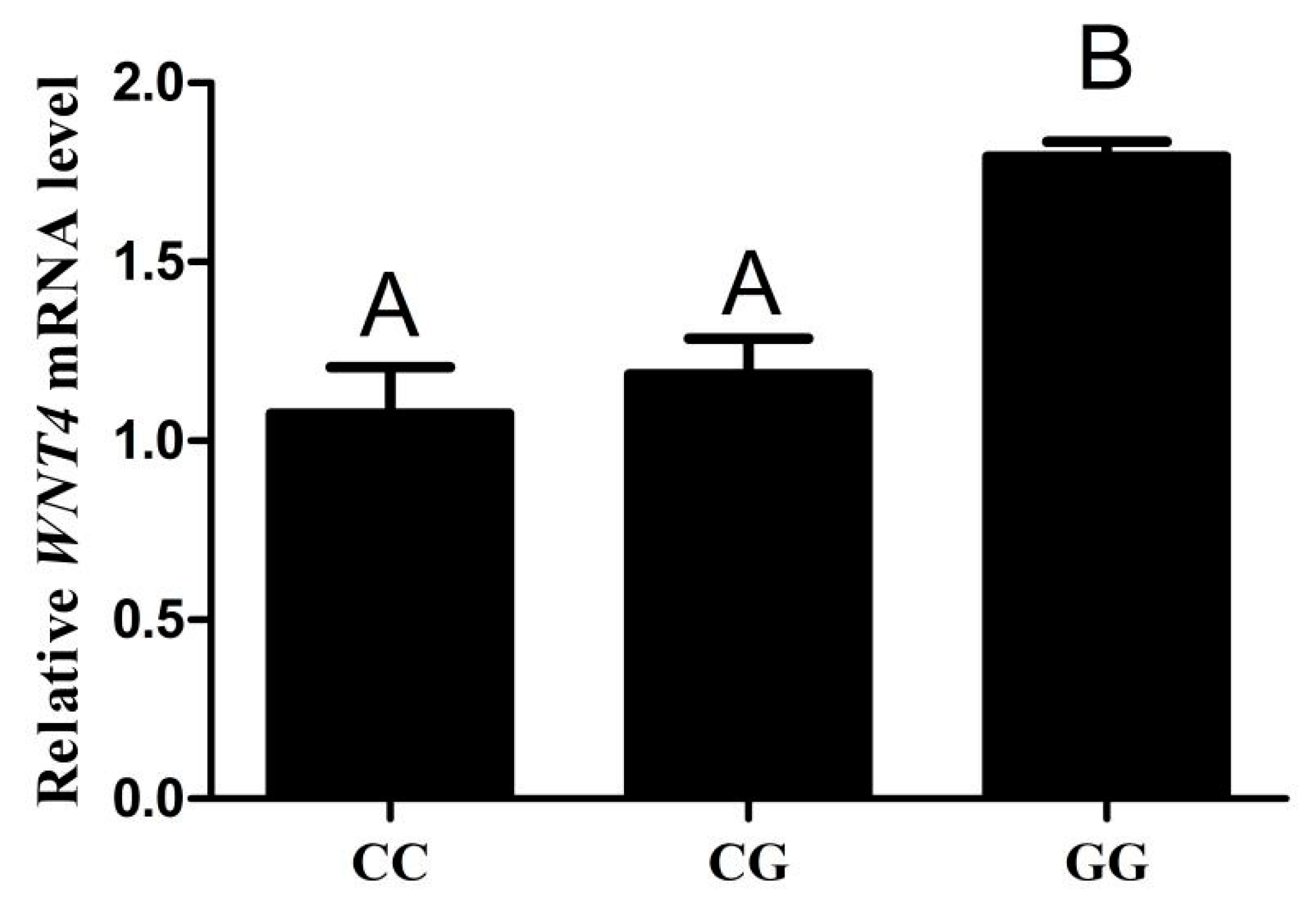
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| pGL3-WNT4(−3354–+252) (F) | CGGGGTACCGCCTGGAGGTATAATAAGCA |
| pGL3-WNT4(−2689–+252) (F) | CGGGGTACCTTAGGTGACGGCACAGCA |
| pGL3-WNT4(−1875–+252) (F) | CGGGGTACCCCAGGGGCATCAGAAT |
| pGL3-WNT4(−1188–+252) (F) | CGGGGTACCTGCGAGCAGGAAAATGGA |
| pGL3-WNT4(−535–+252) (F) | CGGGGTACCGTCTACCAGCAAGAGCAGG |
| Pro-WNT4-1R2 | CCCAAGCTTAGAAGGTGGCGAGGATG |
| WNT4-mut-F | CACATTCCTTTATCCCACGGAAACCTTCACAGTGCA |
| WNT4-mut-R | TGCACTGTGAAGGTTTCCGTGGGATAAAGGAATGTG |
| GAPDH-F | GAGGGTAGTGAAGGCTGCTG |
| GAPDH-R | CACAACACGGTTGCTGTATC |
| WNT4-F | CCTGTCTTTGGCAAGGTGG |
| WNT4-R | CATAGGCAATGTTATCGGAGC |
| Site | Breed (Number) | Genotype Frequency | Allele Frequency | HWE (p-Value) | |||
|---|---|---|---|---|---|---|---|
| g.−3015 | Jining Bairi (n = 539) | GG | CG | CC | G | C | 0.204 |
| 0.358 (n = 193) | 0.458 (n = 247) | 0.184 (n = 99) | 0.587 | 0.413 | |||
| Site | Traits | Genotype | p-Value | ||
|---|---|---|---|---|---|
| g.−3015 | GG (n = 193) | CG (n = 247) | CC (n = 99) | ||
| AFE | 149.763 + 0.411 | 150.518 + 0.409 | 150.235 + 0.764 | 0.473 | |
| E30 | 111.602 ± 1.690 | 113.539 ± 1.074 | 113.083 ± 1.346 | 0.411 | |
| E50 | 137.520 ± 2.039 | 140.223 ± 1.293 | 138.818 ± 1.702 | 0.075 | |
| CLFE | 4.005 ± 0.052 a | 4.080 ± 0.048 ab | 4.224 ± 0.080 b | 0.019 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, C.; Wang, Y.; Liu, C.; Jiang, Y.; Kang, L. A Novel Single-Nucleotide Polymorphism in WNT4 Promoter Affects Its Transcription and Response to FSH in Chicken Follicles. Genes 2022, 13, 1774. https://doi.org/10.3390/genes13101774
Zhong C, Wang Y, Liu C, Jiang Y, Kang L. A Novel Single-Nucleotide Polymorphism in WNT4 Promoter Affects Its Transcription and Response to FSH in Chicken Follicles. Genes. 2022; 13(10):1774. https://doi.org/10.3390/genes13101774
Chicago/Turabian StyleZhong, Conghao, Yiya Wang, Cuiping Liu, Yunliang Jiang, and Li Kang. 2022. "A Novel Single-Nucleotide Polymorphism in WNT4 Promoter Affects Its Transcription and Response to FSH in Chicken Follicles" Genes 13, no. 10: 1774. https://doi.org/10.3390/genes13101774






