QTL Mapping of Stem Rust Resistance in Populations of Durum Wheat
Abstract
1. Introduction
2. Results
2.1. Evaluation of Four Segregating Populations with Their Parents for Reaction to Stem Rust
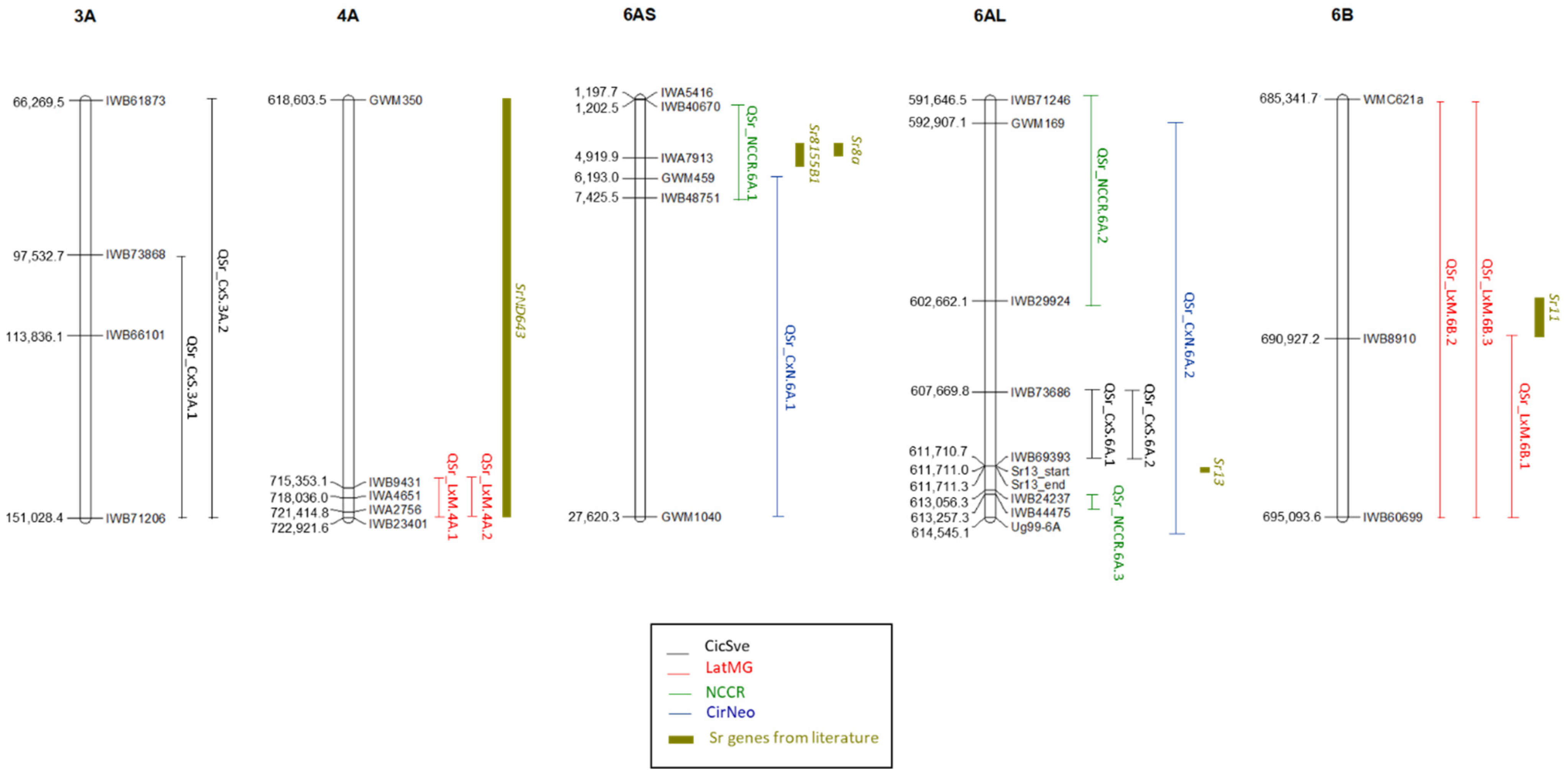
2.2. QTL and Haplotype Analysis
2.3. Identification of Candidate Genes in the QTL Intervals
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Races of the Stem Rust Pathogen
4.2. Phenotypic Evaluation
4.3. Genetic Materials
4.4. Statistical and QTL Analyses
4.5. Identification of Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xynias, I.N.; Mylonas, I.; Korpetis, E.G.; Ninou, E.; Tsaballa, A.; Avdikos, I.D.; Mavromatis, A.G. Durum wheat breeding in the Mediterranean region: Current status and future prospects. Agronomy 2020, 10, 432. [Google Scholar] [CrossRef]
- McVey, D.V.; Long, D.L.; Roberts, J.J. Races of Puccinia graminis in the United States during 1997 and 1998. Plant Dis. 2002, 86, 568–572. [Google Scholar] [CrossRef]
- Jin, Y.; Singh, R.P.; Ward, R.W.; Wanyera, R.; Kinyua, M.; Njau, P.; Pretorius, Z.A. Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 2007, 91, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Szabo, L.J.; Pretorius, Z.A.; Singh, R.P.; Ward, R.; Fetch, T. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 2008, 92, 923–926. [Google Scholar] [CrossRef]
- Rouse, M.N.; Nirmala, J.; Pretorius, Z.A.; Hiebert, C.W. Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor. Appl. Genet. 2014, 127, 1681–1688. [Google Scholar] [CrossRef] [PubMed]
- Patpour, M.; Hovmøller, M.S.; Justesen, A.F.; Newcomb, M.; Olivera, P.; Jin, Y.; Szabo, L.J.; Hodson, D.; Shahin, A.A.; Wanyera, R.; et al. Emergence of virulence to SrTmp in the Ug99 race Group of Wheat Stem Rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis. 2016, 100, 522. [Google Scholar] [CrossRef]
- Olivera, P.D.; Jin, Y.; Rouse, M.; Badebo, A.; Fetch, T.; Singh, R.P.; Yahyaoui, A.M. Races of Puccinia graminis f. sp. tritici with combined virulence to Sr13 and Sr9e in a field stem rust screening nursery in Ethiopia. Plant Dis. 2012, 96, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Olivera, P.D.; Newcomb, M.; Szabo, L.J.; Rouse, M.; Johnson, J.; Gale, S.; Luster, D.G.; Hodson, D.; Cox, J.A.; Burgin, L.; et al. Phenotypic and Genotypic characterization of Race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013–2014. Phytopathology 2015, 105, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Persoons, A.; Bebber, D.P.; Kigathi, R.N.; Maintz, J.; Findlay, K.; Bueno-Sancho, V.; Corredor-Moreno, P.; Harrington, S.A.; Kangara, N.; et al. Potential for re-emergence of wheat stem rust in the United Kingdom. Commun. Biol. 2018, 1, 13. [Google Scholar] [CrossRef]
- Terefe, T.; Pretorius, Z.A.; Visser, B.; Boshoff, W.H.P. First report of Puccinia graminis f. sp. tritici race PTKSK, a variant of wheat stem rust race Ug99 in South Africa. Plant Dis. 2019, 103, 1421. [Google Scholar] [CrossRef]
- Prank, M.; Kenaley, S.C.; Bergstrom, G.C.; Acevedo, M.; Mahowald, N.M. Climate change impacts the spread potential of wheat stem rust, a significant crop disease. Environ. Res. Lett. 2019, 14, 124053. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Jin, Y.; Lagudah, E.S.; Ayliffe, M.A.; Bhavani, S.; Rouse, M.N.; Pretorius, Z.A.; Szabo, L.J.; Huerta-Espino, J.; et al. Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology 2015, 105, 872–884. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.; Morris, C.; Appels, R.; Xia, X. Catalogue of gene symbols for wheat: 2015–2016 supplement. Ann. Wheat Newsl. 2016, 58, 1–18. [Google Scholar]
- Saintenac, C.; Zhang, W.; Salcedo, A.; Rousse, M.; Trick, H.; Akhunov, E.; Dubcovsky, J. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 2013, 341, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Periyannan, S.; Moore, J.; Ayliffe, M.; Bansal, U.; Wang, X.; Huang, L.; Deal, K.; Luo, M.C.; Kong, X.; Bariana, H.; et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 2013, 341, 786–789. [Google Scholar] [CrossRef]
- Mago, R.; Zhang, P.; Vautrin, S.; Simkova, H.; Bansal, U.; Luo, M.-C.; Rouse, M.; Karaoglu, H.; Periyannan, S.; Kolmer, J. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants 2015, 1, 15186. [Google Scholar] [CrossRef]
- Steuernagel, B.; Periyannan, S.K.; Hernandez-Pinzon, I.; Witek, K.; Rouse, M.N.; Yu, G.; Hatta, A.; Ayliffe, M.; Bariana, H.; Jones, J.D.; et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 2016, 34, 652–655. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, S.; Abate, Z.; Nirmala, J.; Rouse, M.N.; Dubcovsky, J. Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Nat. Acad. Sci. USA 2017, 114, E9483–E9492. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, W.; Bolus, S.; Rouse, M.N.; Dubcovsky, J. Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet. 2018, 14, e1007287. [Google Scholar] [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [Google Scholar] [CrossRef]
- Chen, S.; Rouse, M.N.; Zhang, W.; Zhang, X.; Guo, Y.; Briggs, J.; Dubcovsky, J. Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 2020, 225, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Barbier, H.; Rouse, M.N.; Singh, S.; Singh, R.P.; Bhavani, S.; Huerta-Espino, J.; Sorrells, M.E. A consensus map for Ug99 stem rust resistance loci in wheat. Theor. Appl. Genet. 2014, 127, 1561–1581. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Foessel, S.A.; Singh, R.P.; Lillemo, M.; Huerta-Espino, J.; Bhavani, S.; Singh, S.; Lan, C.; Calvo-Salazar, V.; Lagudah, E.S. Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 2014, 127, 781–789. [Google Scholar] [CrossRef]
- Singh, R.P.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Bariana, H.; Bansal, U.; McCallum, B.; Hiebert, C.W.; Bhavani, S.; Singh, S.; Lan, C.; et al. Lr34/Yr18/Sr57/Pm38/Bdv1/Ltn1 confers slow rusting, adult plant resistance to Puccinia graminis tritici. In Proceedings of the 13th International Cereal Rusts and Powdery Mildews Conference, Beijing, China, 28–29 August 2012. [Google Scholar]
- Singh, R.P.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Lan, C.X.; Basnet, B.R.; Bhavani, S. Pleiotropic gene Lr46/Yr29/Pm39/Ltn2 confers slow rusting, adult plant resistance to wheat stem rust fungus. In Proceedings of the 2013 Borlaug Global Rust Initiative Technical Workshop, New Delhi, India, 19–22 August 2013. [Google Scholar]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Hiebert, C.W.; Moscou, M.J.; Hewitt, T.; Steuernagel, B.; Hernández-Pinzón, I.; Green, P.; Pujol, V.; Zhang, P.; Rouse, M.N.; Jin, Y.; et al. Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nat. Comm. 2020, 11, 1123. [Google Scholar] [CrossRef]
- Chen, J.; Upadhyaya, N.M.; Ortiz, D.; Sperschneider, J.; Li, F.; Bouton, C.; Breen, S.; Dong, C.; Xu, B.; Zhang, X.; et al. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 2017, 358, 1607–1610. [Google Scholar] [CrossRef]
- Salcedo, A.; Rutter, W.; Wang, S.; Akhunova, A.; Bolus, S.; Chao, S.; Anderson, N.; Fernandez De Soto, M.; Rouse, M.; Szabo, L.; et al. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 2017, 358, 1604–1606. [Google Scholar] [CrossRef]
- Chen, S.; Guo, Y.; Briggs, J.; Dubach, F.; Chao, S.; Zhang, W.; Rouse, M.N.; Dubcovsky, J. Mapping and characterization of wheat stem rust resistance genes SrTm5 and Sr60 from Triticum monococcum. Theor. Appl. Genet. 2018, 131, 625–635. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome reveals past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef]
- Mazzucotelli, E.; Sciara, G.; Mastrangelo, A.M.; Desiderio, F.; Xu, S.S.; Faris, J.; Hayden, M.J.; Tricker, P.J.; Ozkan, H.; Echenique, V.; et al. The Global Durum wheat panel: An international platform to identify and exchange beneficial alleles. Front. Plant Sci. 2020, 11, 569905. [Google Scholar] [CrossRef] [PubMed]
- Laidò, G.; Panio, G.; Marone, D.; Russo, M.A.; Ficco, D.B.M.; Giovanniello, V.; Cattivelli, L.; Steffenson, B.; DeVita, P.; Mastrangelo, A.M. Identification of new resistance loci to African stem rust race TTKSK in tetraploid wheats based on linkage and genome-wide association mapping. Front. Plant Sci. 2015, 6, 1033. [Google Scholar] [CrossRef] [PubMed]
- Saccomanno, A.; Matny, O.; Marone, D.; Laidò, G.; Petruzzino, G.; Mazzucotelli, E.; Desiderio, F.; Blanco, A.; Gadaleta, A.; Pecchioni, N.; et al. Genetic mapping of loci for resistance to stem rust in a tetraploid wheat collection. Int. J. Mol. Sci. 2018, 19, 3907. [Google Scholar] [CrossRef] [PubMed]
- Hiebert, C.W.; Rouse, M.N.; Nirmala, J.; Fetch, T. Genetic mapping of stem rust resistance to Puccinia graminis f. sp. tritici race TRTTF in the Canadian wheat cultivar Harvest. Phytopathology 2017, 107, 192–197. [Google Scholar] [CrossRef]
- Babiker, E.M.; Gordon, T.C.; Bonman, J.M.; Chao, S.; Rouse, M.N.; Jin, Y.; Newcomb, M.; Wanyera, R.; Bhavani, S. Genetic loci conditioning adult plant resistance to the Ug99 race group and seedling resistance to races TRTTF and TTTTF of the stem rust pathogen in wheat landrace CItr 15026. Plant Dis. 2017, 101, 3. [Google Scholar] [CrossRef]
- Milner, S.G.; Maccaferri, M.; Huang, B.E.; Mantovani, P.; Massi, A.; Frascaroli, E.; Tuberosa, R.; Salvi, S. A multiparental cross population for mapping QTL for agronomic traits in durum wheat (Triticum turgidum ssp. durum). Plant Biotechnol. J. 2016, 14, 735–748. [Google Scholar] [CrossRef]
- Haile, J.K.; Nachit, M.M.; Hammer, K.; Badebo, A.; Roder, M.S. QTL mapping of resistance to race Ug99 of Puccinia graminis f. sp. tritici in durum wheat (Triticum durum Desf.). Mol. Breed. 2012, 30, 1479–1493. [Google Scholar] [CrossRef]
- Letta, T.; Maccaferri, M.; Badebo, A.; Ammar, K.; Ricci, A.; Crossa, J.; Tuberosa, R. Searching for novel sources of field resistance to Ug99 and Ethiopian stem rust races in durum wheat via association mapping. Theor. Appl. Genet. 2013, 126, 1237–1256. [Google Scholar] [CrossRef]
- Mastrangelo, A.M.; Cattivelli, L. What makes bread and durum wheat different? Trends Plant Sci. 2021, 26, 677–684. [Google Scholar] [CrossRef]
- Letta, T.; Olivera, P.; Maccaferri, M.; Jin, Y.; Ammar, K.; Badebo, A.; Salvi, S.; Noli, E.; Crossa, J.; Tuberosa, R. Association mapping reveals novel stem rust resistance loci in durum wheat at the seedling stage. Plant Genome 2014, 7, 1–13. [Google Scholar] [CrossRef]
- Megerssa, S.H.; Ammar, K.; Acevedo, M.; Brown-Guedira, G.; Ward, B.; Degete, A.G.; Randhawa, M.S.; Sorrells, M.E. Multiple-race stem rust resistance loci identified in durum wheat using genome-wide association mapping. Front. Plant Sci. 2020, 11, 598509. [Google Scholar] [CrossRef]
- Saini, D.K.; Srivastava, P.; Pal, N.; Gupta, P.K. Meta-QTLs, ortho-meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2022, 135, 1049–1081. [Google Scholar] [CrossRef]
- Nirmala, J.; Saini, J.; Newcomb, M.; Olivera, P.; Gale, S.; Klindworth, D.; Elias, E.; Talbert, L.; Chao, S.; Faris, J.; et al. Discovery of a novel stem rust resistance allele in durum wheat that exhibits differential reactions to Ug99 isolates. G3 Genes Genomes Genet. 2017, 7, 3481–3490. [Google Scholar] [CrossRef]
- Kosgey, Z.C.; Edae, E.A.; Dill-Macky, R.; Jin, Y.; Bulbula, W.D.; Gemechu, A.; Macharia, G.; Bhavani, S.; Randhawa, M.S.; Rouse, M.N. Mapping and validation of stem rust resistance loci in spring wheat line CI 14275. Front. Plant Sci. 2021, 11, 609659. [Google Scholar] [CrossRef]
- Simons, K.; Abate, Z.; Chao, S.; Zhang, W.; Rouse, M.; Jin, Y.; Elias, E.; Dubcovsky, J. Genetic mapping of stem rust resistance gene Sr13 in tetraploid wheat (Triticum turgidum ssp. durum L.). Theor. Appl. Genet. 2011, 122, 649–658. [Google Scholar] [CrossRef]
- Gill, B.K.; Klindworth, D.L.; Rouse, M.N.; Zhang, J.; Zhang, Q.; Sharma, J.S.; Chu, C.; Long, Y.; Chao, S.; Olivera, P.D.; et al. Function and evolution of allelic variations of Sr13 conferring resistance to stem rust in tetraploid wheat (Triticum turgidum L.). Plant J. 2021, 106, 1674–1691. [Google Scholar] [CrossRef]
- Nirmala, J.; Chao, S.; Olivera, P.; Babiker, E.M.; Abeyo, B.; Tadesse, Z.; Imtiaz, M.; Talbert, L.; Blake, N.K.; Akhunov, E.; et al. Markers linked to wheat stem rust resistance gene Sr11 effective to Puccinia graminis f. sp. tritici race TKTTF. Phytopathology 2016, 106, 1352–1358. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.A.; Laidò, G.; De Vita, P.; Papa, R.; Blanco, A.; Gadaleta, A.; Rubiales, D.; Mastrangelo, A.M. Genetic basis of qualitative and quantitative resistance to powdery mildew in wheat: From consensus regions to candidate genes. BMC Genom. 2013, 14, 562. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Mazzucotelli, E.; Marone, D.; Crosatti, C.; Michelotti, V.; Valè, G.; Mastrangelo, A.M. Regulation and evolution of NLR genes: A close interconnection for plant immunity. Int. J. Mol. Sci. 2018, 19, E1662. [Google Scholar] [CrossRef]
- Colasuonno, P.; Gadaleta, A.; Giancaspro, A.; Nigro, D.; Giove, S.; Incerti, O.; Mangini, G.; Signorile, A.; Simeone, R.; Blanco, A. Development of a high-density SNP-based linkage map and detection of yellow pigment content QTLs in durum wheat. Mol. Breed. 2014, 34, 1563–1578. [Google Scholar] [CrossRef]
- Marone, D.; Panio, G.; Ficco, D.; Russo, M.A.; De Vita, P.; Papa, R.; Rubiales, D.; Cattivelli, L.; Mastrangelo, A.M. Characterization of wheat DArT markers: Genetic and functional features. Mol. Genet. Genom. 2012, 287, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Desiderio, F.; Guerra, D.; Rubiales, D.; Piarulli, L.; Pasquini, M.; Mastrangelo, A.M.; Simeone, R.; Blanco, A.; Cattivelli, L.; Vale’, G. Identification and mapping of quantitative trait loci for leaf rust resistance derived from a tetraploid wheat Triticum dicoccum accession. Mol. Breed. 2014, 34, 1659–1675. [Google Scholar] [CrossRef]
- Roelfs, A.P.; Martens, J.W. An international system of nomenclature for Puccinia graminis f. sp. tritici. Phytopathology 1988, 78, 525–533. [Google Scholar] [CrossRef]
- Huang, S.; Steffenson, B.J.; Sela, H.; Stinebaugh, K. Resistance of Aegilops longissima to the rusts of wheat. Plant Dis. 2018, 102, 1224–1235. [Google Scholar] [CrossRef]
- Stakman, E.C.; Stewart, D.M.; Loegering, W.Q. Identification of Physiologic Races of Puccinia graminis var. tritici; USDA: Washington, DC, USA, 1962. [Google Scholar]
- Zhang, D.; Bowden, R.L.; Yu, J.; Carver, B.F.; Bai, G. Association analysis of stem rust resistance in US winter wheat. PLoS ONE 2014, 9, e103747. [Google Scholar]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.B. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [CrossRef]
- Blanco, A.; Colasuonno, P.; Gadaleta, A.; Mangini, G.; Schiavulli, A.; Simeone, R.; Digesù, A.M.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L. Quantitative trait loci for yellow pigment concentration and individual carotenoid compounds in durum wheat. J. Cereal Sci. 2011, 54, 255–264. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.; Weisberg, S. An {R} Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019; Available online: https://socialsciences.mcmaster.ca/jfox/Books/Companion/ (accessed on 1 June 2022).
- R Core Team. R: A Language and Environment for Statistical Computing. 2020. Available online: https://www.r-project.org/ (accessed on 1 June 2022).

| RIL Population Name | Stem Rust Race | No. of Resistant RILs (IT ≤ 6) | No. of Susceptible RILs (IT > 6) | Observed Segregation | χ2 | No. of Expected Genes |
|---|---|---|---|---|---|---|
| LatMG | TPKMC | 66 | 47 | 1:1 | (1, N = 113) = 3.2, p > 0.10 | 1 gene |
| LatMG | JRCQC | 82 | 27 | 3:1 | (1, N = 109) = 0.003, p > 0.90 | 2 genes, both from MG5323 |
| LatMG | TKTTF | 85 | 25 | 3:1 | (1, N = 110) = 0.30, p > 0.50 | 2 genes, both from MG5323 |
| CicSve | TPMKC | 36 | 38 | 1:1 | (1, N = 74) = 0.054, p > 0.80 | 1 gene |
| CicSve | TKTTF | 25 | 25 | 1:1 | exact segregation | 1 gene |
| CirNeo | TTTTF | 85 | 26 | 3:1 | (1, N = 111) = 0.147, p > 0.50 | 2 genes, one per parent |
| NCCR | TTTTF | 213 | 122 | 5:3 | (1, N = 335) = 0.167, p > 0.50 | 3 genes: one independent by two parents, and two dependent on one parent |
| Race | Population | Mean | Range | CV | MSD | Genetic Variance | H2 |
|---|---|---|---|---|---|---|---|
| TPMKC | LatMG | 4.33 | 0.0–9.0 | 0.81 | 2.00 | 6.76 | 0.81 |
| JRCQC | LatMG | 3.16 | 0.0–9.0 | 0.98 | 1.40 | 5.75 | 0.89 |
| TKTTF | LatMG | 3.11 | 0.0–9.0 | 0.96 | 2.11 | 4.69 | 0.74 |
| TPMKC | CicSve | 5.21 | 0.0–9.0 | 0.68 | 1.15 | 5.63 | 0.92 |
| TKTTF | CicSve | 5.44 | 3.0–9.0 | 0.59 | 2.00 | 3.96 | 0.73 |
| TTTTF | CirNeo | 2.34 | 0.0–9.0 | 1.53 | 0.76 | 8.23 | 0.98 |
| TTTTF | NCCR | 3.78 | 0.0–9.0 | 0.94 | 0.27 | 8.35 | 0.99 |
| (a) | ||||
|---|---|---|---|---|
| LatMG | JRCQC | TKTTF | ||
| TPMKC | 0.646 | 0.598 | ||
| JRCQC | 0.893 | |||
| (b) | ||||
| CicSve | TKTTF | |||
| TPMKC | 0.946 | |||
| QTL Name | Trait | Population | Peak Marker | Linkage Group | Position (cM) | LOD | R2 | Add. Effect | CI Start (cM) | CI End (cM) | Left Marker | Right Marker | Left Pos. | Right Pos. | Resistant Parent |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QSr_CxS.3A.1 | TKTTF | CicSve | IWB66101 | 3A_2 | 6 | 3.5 | 0.28 | 1.74 | 3.6 | 8.4 | IWB73868 | IWB71206 | 97,532,661 | 151,028,408 | Svevo |
| QSr_CxS.3A.2 | TPMKC | CicSve | IWB66101 | 3A_2 | 6 | 2.2 | 0.13 | 1.28 | 0.8 | 11.2 | IWB61873 | IWB71206 | 66,269,458 | 151,028,408 | Svevo |
| QSr_LxM.4A.1 | JRCQC | LatMG | IWB72220 | 4A | 152.9 | 15.9 | 0.35 | 1.55 | 143.2 | 153.5 | IWB9431 | IWA2756 | 715,353,110 | 721,414,881 | MG5323 |
| QSr_LxM.4A.2 | TKTTF | LatMG | IWB72220 | 4A | 152.9 | 17.1 | 0.42 | 1.56 | 143.2 | 153.5 | IWB9431 | IWA2756 | 715,353,110 | 721,414,881 | MG5323 |
| QSr_NCCR.6A.1 | TTTTF | NCCR | IWB48751 | 6A | 7 | 39.1 | 0.27 | 4.39 | 0 | 7 | IWB40670 | IWB48751 | 1,202,560 | 7,425,521 | Neodur |
| QSr_CxN.6A.1 | TTTTF | CirNeo | Xgwm459 | 6A-1 | 95.8 | 13.3 | 0.42 | 2.286 | 94.2 | 95.8 | Xgwm459 | Xgwm1040 | 6,193,023 | 27,620,384 | Neodur |
| QSr_NCCR.6A.2 | TTTTF | NCCR | IWB29924 | 6A | 116.7 | 54.9 | 0.36 | 4.4 | 10.7 | 116.7 | IWB71246 | IWB29924 | 591,646,505 | 602,662,186 | Claudio-Rascon/2*Tarro |
| QSr_NCCR.6A.3 | TTTTF | NCCR | IWB60184 | 6A | 125.7 | 70.9 | 0.46 | 4.81 | 125.7 | 125.7 | IWB24237 | IWB44475 | 613,056,324 | 613,257,397 | Claudio-Rascon/2*Tarro |
| QSr_CxS.6A.1 | TKTTF | CicSve | IWB69393 | 6A_5 | 4 | 27.9 | 0.92 | 2.99 | 3.3 | 4.7 | IWB73686 | IWB69393 | 607,669,872 | 611,710,729 | Svevo |
| QSr_CxS.6A.2 | TPMKC | CicSve | IWB69393 | 6A_5 | 4 | 40.4 | 0.92 | 3.399 | 3.3 | 4.7 | IWB73686 | IWB69393 | 607,669,872 | 611,710,729 | Svevo |
| QSr_CxN.6A.2 | TTTTF | CirNeo | Ug99-6A | 6A-2 | 0 | 9.9 | 0.34 | 1.763 | 0 | 4 | Xgwm169 | Ug99-6A * | 592,907,113 | 614,545,144 | Cirillo |
| QSr_LxM.6B.1 | TPKMC | LatMG | IWB60699 | 6B | 148.7 | 23.3 | 0.68 | 2.91 | 147.6 | 148.7 | IWB8910 | IWB60699 | 690,927,250 | 695,093,664 | MG5323 |
| QSr_LxM.6B.2 | JRCQC | LatMG | IWB60699 | 6B | 148.7 | 18.5 | 0.44 | 1.77 | 147 | 148.7 | Xwmc621a | IWB60699 | 685,341,777 | 695,093,664 | MG5323 |
| QSr_LxM.6B.3 | TKTTF | LatMG | IWB58435 | 6B | 148.1 | 16.9 | 0.41 | 1.46 | 146.3 | 148.7 | Xwmc621a | IWB60699 | 685,341,777 | 695,093,664 | MG5323 |
| QTL | Interval Svevo (Mbp) | Number of Annotated Genes | Ratio Annotated Genes/Mbp | Number of Disease-Related Genes | Ratio Disease-Related/Annotated Genes | Ratio Disease-Related Genes/Mbp |
|---|---|---|---|---|---|---|
| QSr_CxS.3A.1 | 53.5 | 319 | 6.0 | 33 | 0.1 | 0.6 |
| QSr_CxS.3A.2 | 84.8 | 517 | 6.1 | 47 | 0.1 | 0.5 |
| QSr_LxM.4A.1/2 | 6.1 | 139 | 23.3 | 20 | 0.1 | 3.3 |
| QSr_NCCR.6A.1 | 6.2 | 134 | 21.5 | 22 | 0.2 | 3.5 |
| QSr_CxN.6A.1 | 21.4 | 927 | 43.3 | 116 | 0.1 | 5.4 |
| QSr_NCCR.6A.2 | 11.0 | 712 | 64.6 | 46 | 0.1 | 4.2 |
| QSr_NCCR.6A.3 | 0.2 | 18 | 89.5 | 2 | 0.1 | 10.0 |
| QSr_CxS.6A.1/2 | 4.0 | 46 | 11.4 | 11 | 0.2 | 2.7 |
| QSr_CxN.6A.2 | 21.6 | 1136 | 52.5 | 116 | 0.1 | 5.4 |
| QSr_LxM.6B.1 | 4.2 | 210 | 50.4 | 30 | 0.1 | 7.1 |
| QSr_LxM.6B.2/3 | 9.8 | 422 | 43.3 | 72 | 0.2 | 7.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marone, D.; Mazzucotelli, E.; Matny, O.; Desiderio, F.; Sciara, G.; Maccaferri, M.; Marcotuli, I.; Gadaleta, A.; Steffenson, B.; Mastrangelo, A.M. QTL Mapping of Stem Rust Resistance in Populations of Durum Wheat. Genes 2022, 13, 1793. https://doi.org/10.3390/genes13101793
Marone D, Mazzucotelli E, Matny O, Desiderio F, Sciara G, Maccaferri M, Marcotuli I, Gadaleta A, Steffenson B, Mastrangelo AM. QTL Mapping of Stem Rust Resistance in Populations of Durum Wheat. Genes. 2022; 13(10):1793. https://doi.org/10.3390/genes13101793
Chicago/Turabian StyleMarone, Daniela, Elisabetta Mazzucotelli, Oadi Matny, Francesca Desiderio, Giuseppe Sciara, Marco Maccaferri, Ilaria Marcotuli, Agata Gadaleta, Brian Steffenson, and Anna Maria Mastrangelo. 2022. "QTL Mapping of Stem Rust Resistance in Populations of Durum Wheat" Genes 13, no. 10: 1793. https://doi.org/10.3390/genes13101793
APA StyleMarone, D., Mazzucotelli, E., Matny, O., Desiderio, F., Sciara, G., Maccaferri, M., Marcotuli, I., Gadaleta, A., Steffenson, B., & Mastrangelo, A. M. (2022). QTL Mapping of Stem Rust Resistance in Populations of Durum Wheat. Genes, 13(10), 1793. https://doi.org/10.3390/genes13101793










