Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of PtriZAT Members in P. trichocarpa and Populus Speciecs
2.2. Phylogenetic Relationship of ZAT Proteins in P. trichocarpa
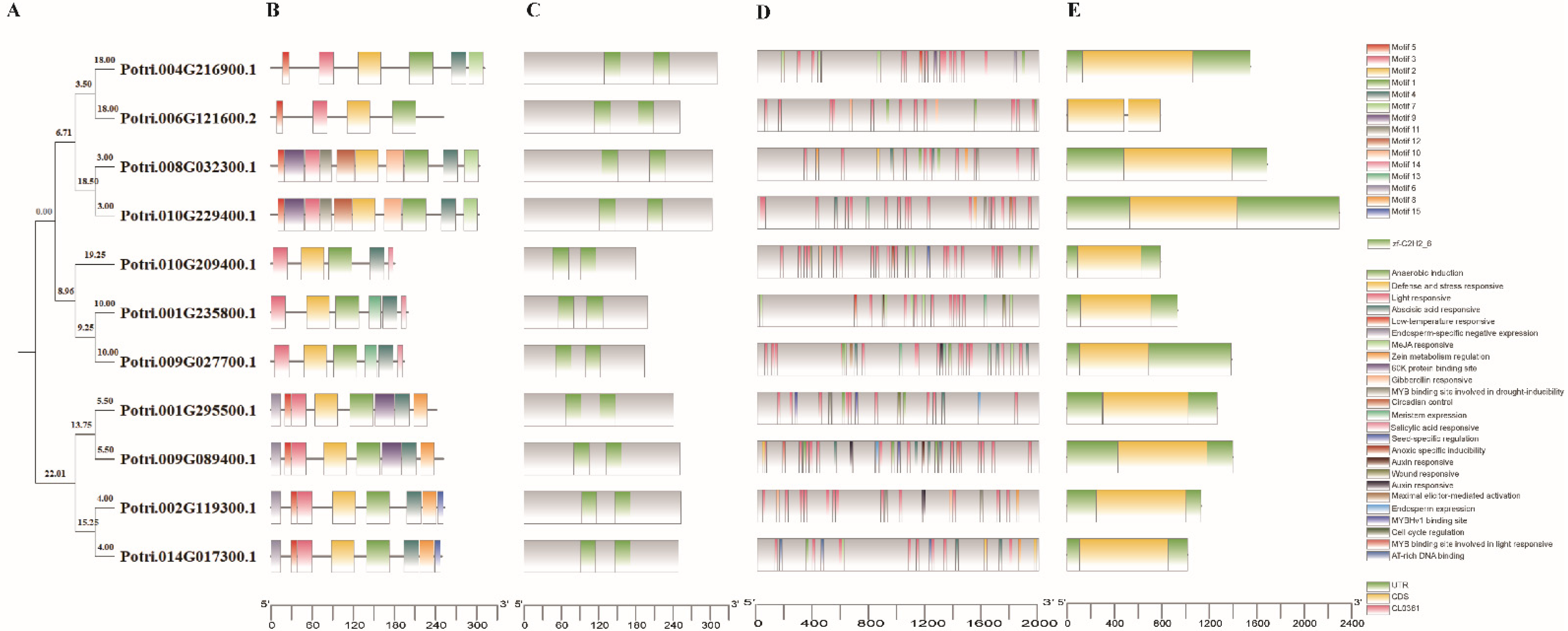
2.3. Exon Intron Structure, Location, Conserved Motifs and Promoter in PtriZAT Genes
2.4. Plant Materials and qRT-PCR Assays
2.5. Expression Profiles of PtriZAT Genes in Different Plants
2.6. Prediction of the Interaction Proteins of PtriZATs
3. Results
3.1. Genome-Wide Identification of PtriZAT Members in P. trichocarpa
3.2. Sequence Characteristics and Phylogenetic Relationships of PtriZATs
3.3. Gene, Protein Structure, and Conserved Motifs of PtriZAT Genes
3.4. Chromosomal Location and Duplication Events of PtriZAT Genes
3.5. Cis-Elements in the Promoter Regions of PtriZAT Genes
3.6. Expression Patterns of PtriZAT Genes in Different Tissues and Stress Treatment
3.7. Protein–Protein Interaction Networks
3.8. The Distribution of PtriZATs in Populus Species
4. Discussion
4.1. Characteristics of PtriZATs in P. trichocarpa
4.2. Different Distributions of ZATs in Populus Species
4.3. Chromosomal Distribution and Gene Duplication of PtriZATs
4.4. Functional Differentiation of PtriZATs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Gamsjaeger, R.; Liew, C.K.; Loughlin, F.E.; Crossley, M.; Mackay, J.P. Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007, 32, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Kant, R.; Pradhan, A.; Jha, G. RS_CRZ1, a C2H2-Type Transcription Factor Is Required for Pathogenesis of Rhizoctonia solani AG1-IA in Tomato. Mol. Plant Microbe Interact. 2021, 34, 26–38. [Google Scholar] [CrossRef]
- Ballerini, E.S.; Min, Y.; Edwards, M.B.; Kramer, E.M.; Hodges, S.A. POPOVICH, encoding a C2H2 zinc-finger transcription factor, plays a central role in the development of a key innovation, floral nectar spurs, in Aquilegia. Proc. Natl. Acad. Sci. USA 2020, 117, 22552–22560. [Google Scholar] [CrossRef]
- Arrey-Salas, O.; Caris-Maldonado, J.C.; Hernández-Rojas, B.; Gonzalez, E. Comprehensive Genome-Wide Exploration of C2H2 Zinc Finger Family in Grapevine (Vitis vinifera L.): Insights into the Roles in the Pollen Development Regulation. Genes 2021, 12, 302. [Google Scholar] [CrossRef]
- Chang, J.; Yu, T.; Yang, Q.; Li, C.; Xiong, C.; Gao, S.; Xie, Q.; Zheng, F.; Li, H.; Tian, Z.; et al. Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018, 96, 90–102. [Google Scholar] [CrossRef] [Green Version]
- Shang, L.; Song, J.; Yu, H.; Wang, X.; Yu, C.; Wang, Y.; Li, F.; Lu, Y.; Wang, T.; Ouyang, B.; et al. A mutation in a C2H2-type zinc finger transcription factor contributed to the transition toward self-pollination in cultivated tomato. Plant Cell 2021, 33, 3293–3308. [Google Scholar] [CrossRef]
- Zhuang, H.; Wang, H.L.; Zhang, T.; Zeng, X.Q.; Chen, H.; Wang, Z.W.; Zhang, J.; Zheng, H.; Tang, J.; Ling, Y.H.; et al. NONSTOP GLUMES1 Encodes a C2H2 Zinc Finger Protein That Regulates Spikelet Development in Rice. Plant Cell 2020, 32, 392–413. [Google Scholar] [CrossRef]
- Ding, W.; Wang, Y.; Fang, W.; Gao, S.; Li, X.; Xiao, K. TaZAT8, a C2H2-ZFP type transcription factor gene in wheat, plays critical roles in mediating tolerance to Pi deprivation through regulating P acquisition, ROS homeostasis and root system establishment. Physiol. Plant 2016, 158, 297–311. [Google Scholar] [CrossRef]
- Cheuk, A.; Ouellet, F.; Houde, M. The barley stripe mosaic virus expression system reveals the wheat C2H2 zinc finger protein TaZFP1B as a key regulator of drought tolerance. BMC Plant Biol. 2020, 20, 144. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Li, H.G.; Wang, J.J.; Su, Y.; Wang, H.L.; Feng, C.H.; Yang, Y.; Niu, M.X.; Liu, C.; Yin, W.; et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 2019, 17, 2169–2183. [Google Scholar] [CrossRef] [Green Version]
- Kundu, A.; Das, S.; Basu, S.; Kobayashi, Y.; Kobayashi, Y.; Koyama, H.; Ganesan, M. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019, 21, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Yan, H.; Liang, L.; Zhang, Y.; Yang, H.; Li, W.; Choi, J.; Huang, J.; Deng, S. A C2H2-Type Zinc-Finger Protein from Millettia pinnata, MpZFP1, Enhances Salt Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10832. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, J.; Zhong, G.; Wang, B. Genome-Wide Identification and Expression Patterns of the C2H2-Zinc Finger Gene Family Related to Stress Responses and Catechins Accumulation in C. sinensis [L.] O. Kuntze. Int. J. Mol. Sci. 2021, 22, 4197. [Google Scholar] [CrossRef]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The Roles of Arabidopsis C1-2i Subclass of C2H2-type Zinc-Finger Transcription Factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef] [Green Version]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the A. thaliana genome. BMC Genom. 2004, 5, 39. [Google Scholar]
- Sakamoto, H.; Araki, T.; Meshi, T.; Iwabuchi, M. Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress. Gene 2000, 248, 23–32. [Google Scholar] [CrossRef]
- Van der Zaal, B.J.; Neuteboom, L.W.; Pinas, J.E.; Chardonnens, A.N.; Schat, H.; Verkleij, J.A.; Hooykaas, P.J. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999, 119, 1047–1055. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Liu, R.; Guo, B.; Huang, K.; Wang, L.; Han, Y.; Li, H.; Hou, S. Ectopic expression of GmZAT4, a putative C2H2-type zinc finger protein, enhances PEG and NaCl stress tolerances in A. thaliana. 3 Biotech 2019, 9, 166. [Google Scholar] [CrossRef]
- Yang, K.; Li, C.Y.; An, J.P.; Wang, D.R.; Wang, X.; Wang, C.K.; You, C.X. The C2H2-type zinc finger transcription factor MdZAT10 negatively regulates drought tolerance in apple. Plant Physiol. Biochem. 2021, 167, 390–399. [Google Scholar] [CrossRef]
- Xing, L.; Qi, S.; Zhou, H.; Zhang, W.; Zhang, C.; Ma, W.; Zhang, Q.; Shah, K.; Han, M.; Zhao, J. Epigenomic Regulatory Mechanism in Vegetative Phase Transition of M. hupehensis. J. Agric. Food Chem. 2020, 68, 4812–4829. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Regier, N.; Frey, B. Experimental comparison of relative RT-qPCR quantification approaches for gene expression studies in poplar. BMC Mol. Biol. 2010, 11, 57. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, 111988. [Google Scholar] [CrossRef]
- Tippmann, H.F. Analysis for free: Comparing programs for sequence analysis. Brief Bioinform. 2004, 5, 82–87. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, P. trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Zolotarev, N.; Fedotova, A.; Kyrchanova, O.; Bonchuk, A.; Penin, A.A.; Lando, A.S.; Eliseeva, I.A.; Kulakovskiy, I.V.; Maksimenko, O.; Georgiev, P. Architectural proteins Pita, Zw5, and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 2016, 44, 7228–7241. [Google Scholar] [PubMed] [Green Version]
- Singh, A.K.; Saharan, K.; Baral, S.; Vasudevan, D. The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction. Biochim. Biophys. Acta Gene Regul. Mech. 2022, 1865, 194872. [Google Scholar] [CrossRef] [PubMed]
- Artegiani, B.; Labbaye, C.; Sferra, A.; Quaranta, M.T.; Torreri, P.; Macchia, G.; Ceccarini, M.; Petrucci, T.C.; Macioce, P. The interaction with HMG20a/b proteins suggests a potential role for β-dystrobrevin in neuronal differentiation. J. Biol. Chem. 2010, 285, 24740–24750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezquita, J.; Chiva, M.; Vidal, S.; Mezquita, C. Effect of high mobility group nonhistone proteins HMG-20 (ubiquitin) and HMG-17 on histone deacetylase activity assayed in vitro. Nucleic Acids Res. 1982, 10, 1781–1797. [Google Scholar] [CrossRef]
- Wang, Y.; Appiah-Kubi, K.; Lan, T.; Wu, M.; Pang, J.; Qian, H.; Tao, Y.; Jiang, L.; Wu, Y.; Chen, Y. PKG II inhibits PDGF-BB triggered biological activities by phosphorylating PDGFRβ in gastric cancer cells. Cell Biol. Int. 2018, 42, 1358–1369. [Google Scholar] [CrossRef]
- Qian, H.; Tao, Y.; Jiang, L.; Wang, Y.; Lan, T.; Wu, M.; Pang, J.; Appiah-Kubi, K.; Chen, Y.; Wu, Y. PKG II effectively reversed EGF-induced protein expression alterations in human gastric cancer cell lines. Cell Biol. Int. 2018, 42, 435–442. [Google Scholar] [CrossRef]
- Wu, Y.; Cai, Q.; Li, W.; Cai, Z.; Liu, Y.; Li, H.; Pang, J.; Chen, Y. Active PKG II inhibited the growth and migration of ovarian cancer cells through blocking Raf/MEK and PI3K/Akt signaling pathways. Biosci. Rep. 2019, 39, BSR20190405. [Google Scholar] [CrossRef] [Green Version]
- Habu, Y. Epigenetic silencing of endogenous repetitive sequences by MORPHEUS’ MOLECULE1 in A. thaliana. Epigenetics 2010, 5, 562–565. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Sun, G.; Shi, C.; Sun, D. Transcriptome analysis reveals new microRNAs-mediated pathway involved in anther development in male sterile wheat. BMC Genom. 2018, 19, 333. [Google Scholar] [CrossRef] [Green Version]
- Galvão, R.M.; Kota, U.; Soderblom, E.J.; Goshe, M.B.; Boss, W.F. Characterization of a new family of protein kinases from Arabidopsis containing phosphoinositide 3/4-kinase and ubiquitin-like domains. Biochem. J. 2008, 409, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Kubo, K.; Sakamoto, A.; Kobayashi, A.; Rybka, Z.; Kanno, Y.; Nakagawa, H.; Takatsuji, H. Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998, 26, 608–715. [Google Scholar] [CrossRef] [Green Version]
- Enuameh, M.S.; Asriyan, Y.; Richards, A.; Christensen, R.G.; Hall, V.L.; Kazemian, M.; Zhu, C.; Pham, H.; Cheng, Q.; Blatti, C.; et al. Global analysis of Drosophila Cys₂-His₂ zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013, 23, 928–940. [Google Scholar] [CrossRef]
- Persikov, A.V.; Wetzel, J.L.; Rowland, E.F.; Oakes, B.L.; Xu, D.J.; Singh, M.; Noyes, M.B. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015, 43, 1965–1984. [Google Scholar] [CrossRef] [Green Version]
- Meseguer, A.; Årman, F.; Fornes, O.; Molina-Fernández, R.; Bonet, J.; Fernandez-Fuentes, N.; Oliva, B. On the prediction of DNA-binding preferences of C2H2-ZF domains using structural models: Application on human CTCF. NAR Genom. Bioinform. 2020, 2, 46. [Google Scholar] [CrossRef]
- Najafabadi, H.S.; Garton, M.; Weirauch, M.T.; Mnaimneh, S.; Yang, A.; Kim, P.M.; Hughes, T.R. Non-base-contacting residues enable kaleidoscopic evolution of metazoan C2H2 zinc finger DNA binding. Genome Biol. 2017, 18, 167. [Google Scholar] [CrossRef] [Green Version]
- Gill, G.; Pascal, E.; Tseng, Z.H.; Tjian, R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 1994, 91, 192–196. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, Z.; Xu, X.; Zhang, H.; Li, C. Genome-Wide Analysis of C2H2 Zinc-Finger Family Transcription Factors and Their Responses to Abiotic Stresses in Poplar (P. trichocarpa). PLoS ONE 2015, 10, e0134753. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Yang, Y.; Wang, Z.; Zhao, P.; Yuan, Y.; Zhang, L.; Ma, Y.; Pang, C.; Yu, J.; Xiao, G. Comprehensive analyses of ZFP gene family and characterization of expression profiles during plant hormone response in cotton. BMC Plant Biol. 2019, 19, 329. [Google Scholar] [CrossRef] [Green Version]
- Martin, L.; Leblanc-Fournier, N.; Azri, W.; Lenne, C.; Henry, C.; Coutand, C.; Julien, J.-L. Characterization and expression analysis under bending and other abiotic factors of PtaZFP2, a poplar gene encoding a Cys2/His2 zinc finger protein. Tree Physiol. 2009, 29, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhang, S.; Zhao, H.; Korpelainen, H.; Li, C. Sex-related adaptive responses to interaction of drought and salinity in Populus yunnanensis. Plant Cell Environ. 2010, 33, 1767–1778. [Google Scholar] [CrossRef]
- Melnikova, N.V.; Borkhert, E.V.; Snezhkina, A.V.; Kudryavtseva, A.V.; Dmitriev, A.A. Sex-Specific Response to Stress in Populus. Front. Plant Sci. 2017, 8, 1827. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Chen, L.; Zhu, P.; Zhang, J.; Zhang, D.; Xiao, J.; Xu, Z.; Zhang, L.; Liu, Y.; Li, H.; et al. Sex-specific responses of Populus deltoides to interaction of cadmium and salinity in root systems. Ecotoxicol. Environ. Saf. 2020, 195, 110437. [Google Scholar] [CrossRef]
- Gorrec, F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 2009, 42, 1035–1042. [Google Scholar] [CrossRef]
- Xu, X.; Qiao, D.; Dong, C.; Mann, M.; Garofalo, R.P.; Keles, S.; Brasier, A.R. The SWI/SNF-Related, Matrix Associated, Actin-Dependent Regulator of Chromatin A4 Core Complex Represses Respiratory Syncytial Virus-Induced Syncytia Formation and Subepithelial Myofibroblast Transition. Front. Immunol. 2021, 12, 633654. [Google Scholar] [CrossRef]






| ID. | Number of Amino Acids | Zn-C2H2 Domain1 | Zn-C2H2 Domain2 | Molecular Weight | Theoretical pI | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Location |
|---|---|---|---|---|---|---|---|---|
| Potri.001G235800.1.p | 198 | 55–80 | 101–127 | 21,440.73 | 9.62 | 77.02 | −0.399 | nucle- |
| Potri.002G119300.1.p | 252 | 93–117 | 146–171 | 27,162.32 | 7.7 | 59.68 | −0.57 | nucle- |
| Potri.004G216900.1.p | 310 | 129–155 | 209–234 | 33,781.64 | 7.23 | 62.03 | −0.695 | nucle- |
| Potri.006G121600.2.p | 250 | 184–209 | 113–139 | 27,202.53 | 9.01 | 63.24 | −0.63 | nucle- |
| Potri.008G032300.1.p | 303 | 125–151 | 202–228 | 33,762.19 | 7.74 | 55.38 | −0.938 | nucle- |
| Potri.009G027700.1.p | 193 | 51–76 | 99–123 | 21,035.39 | 9.57 | 76.89 | −0.41 | nucle- |
| Potri.009G089400.1.p | 250 | 80–105 | 132–157 | 26,305.14 | 8.83 | 55.12 | −0.657 | nucle- |
| Potri.010G209400.1.p | 179 | 46–72 | 91–116 | 20,059.96 | 9.06 | 61.62 | −0.523 | nucle- |
| Potri.010G229400.1.p | 302 | 121–147 | 199–223 | 33,286.63 | 7.21 | 55.93 | −0.901 | nucle- |
| Potri.014G017300.1.p | 248 | 91–115 | 146–171 | 26,605.77 | 8.4 | 58.27 | −0.56 | nucle- |
| Potri.001G295500.1.p | 240 | 67–92 | 122–147 | 25,363.26 | 8.45 | 56.62 | −0.577 | nucle- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Yu, A.; Sun, R.; Liu, A. Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR. Genes 2022, 13, 1843. https://doi.org/10.3390/genes13101843
Li P, Yu A, Sun R, Liu A. Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR. Genes. 2022; 13(10):1843. https://doi.org/10.3390/genes13101843
Chicago/Turabian StyleLi, Ping, Anmin Yu, Rui Sun, and Aizhong Liu. 2022. "Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR" Genes 13, no. 10: 1843. https://doi.org/10.3390/genes13101843
APA StyleLi, P., Yu, A., Sun, R., & Liu, A. (2022). Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR. Genes, 13(10), 1843. https://doi.org/10.3390/genes13101843







