ACADM Frameshift Variant in Cavalier King Charles Spaniels with Medium-Chain Acyl-CoA Dehydrogenase Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Examination and Investigations
2.2. DNA Extraction
2.3. Whole-Genome Sequencing
2.4. Variant Calling
2.5. Gene Analysis
2.6. Allele Specific PCR and Sanger Sequencing
2.7. Acylcarnitine Screening
3. Results
3.1. Clinical History, Examination and Investigations
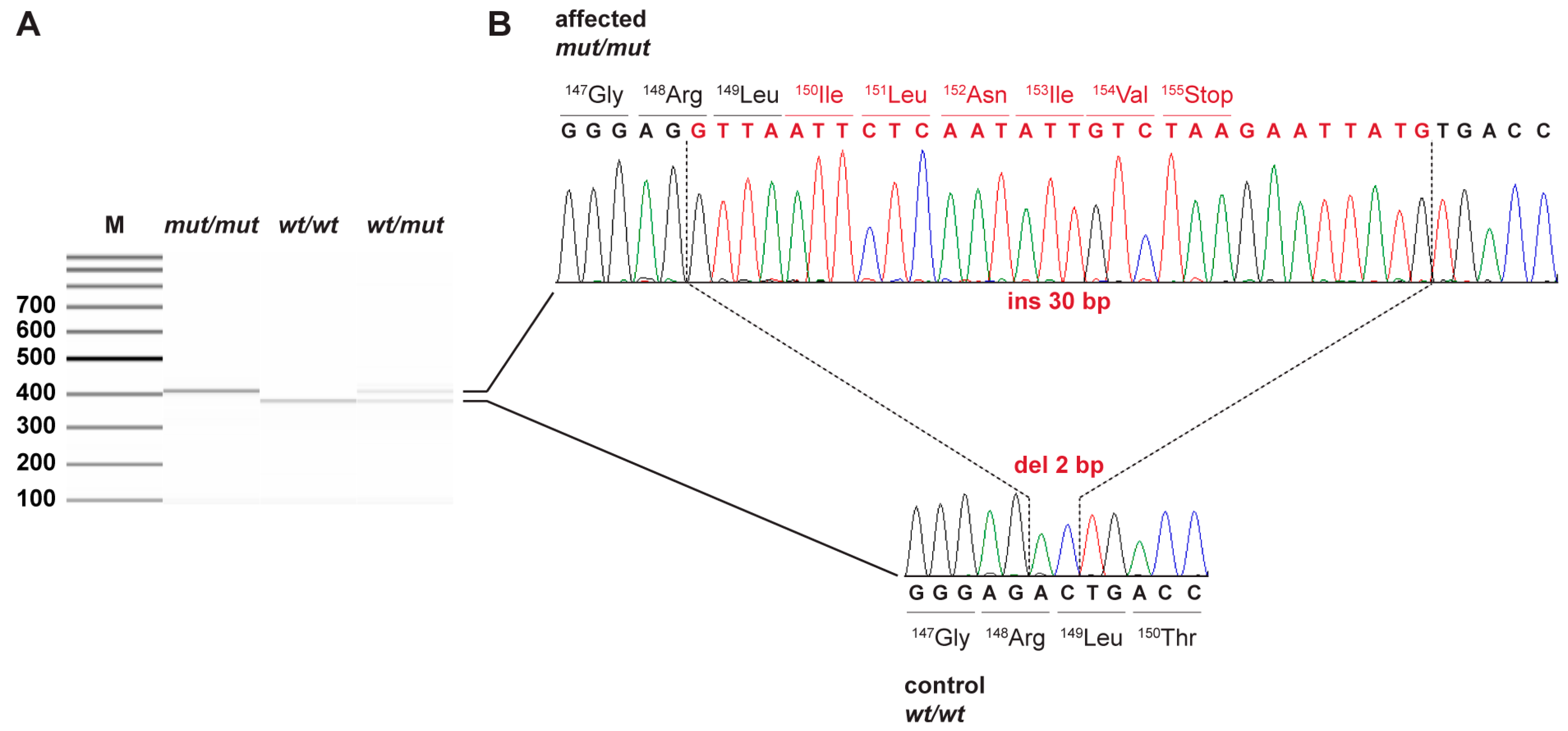
3.2. Genetic Analysis
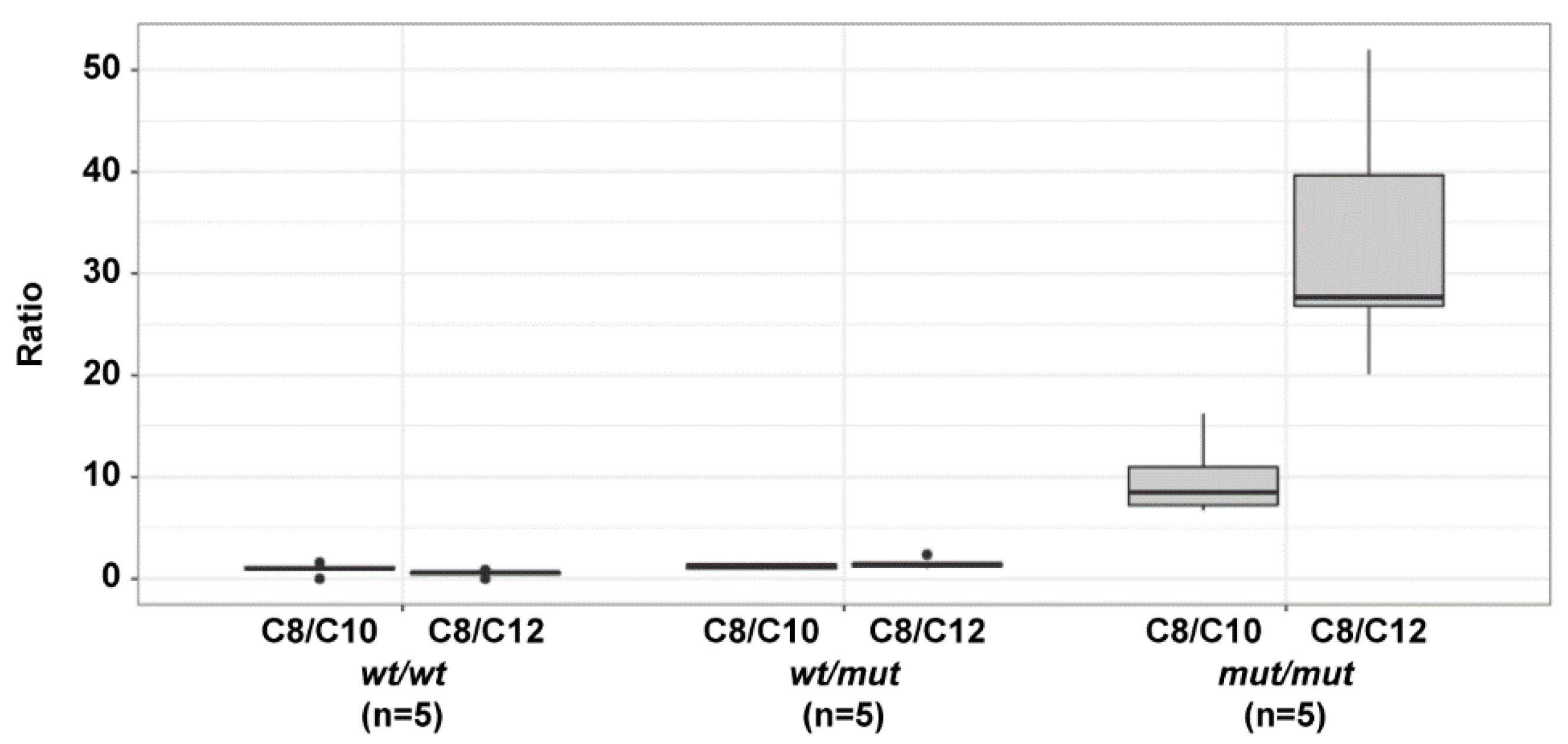
3.3. Acylcarnitine Measurements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vishwanath, V.A. Fatty Acid Beta-Oxidation Disorders: A Brief Review. Ann. Neurosci. 2016, 23, 51–55. [Google Scholar] [CrossRef] [PubMed]
- van Beusekom, C.; Martini, I.A.; Rutgers, H.M.; Boersma, E.R.; Muskiet, F.A. A carbohydrate-rich diet not only leads to incorporation of medium-chain fatty acids (6:0-14:0) in milk triglycerides but also in each milk-phospholipid subclass. Am. J. Clin. Nutr. 1990, 52, 326–334. [Google Scholar] [CrossRef]
- Leslie, N.D.; Saenz-Ayala, S. Very Long-Chain Acyl-Coenzyme a Dehydrogenase Deficiency; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6816/ (accessed on 21 September 2022).
- Schönfeld, P.; Wojtczak, L. Short- and medium-chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef]
- Andresen, B.S.; Dobrowolski, S.F.; O’Reilly, L.; Muenzer, J.; McCandless, S.E.; Frazier, D.M.; Udvari, S.; Bross, P.; Knudsen, I.; Banas, R.; et al. Medium-chain acyl-CoA dehydrogenase (MCAD) mutations identified by MS/MS-based prospective screening of newborns differ from those observed in patients with clinical symptoms: Identification and characterization of a new, prevalent mutation that results in mild MCAD deficiency*. Am. J. Hum. Genet. 2001, 68, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Spiekerkoetter, U.; Duran, M. Mitochondrial fatty acid oxidation disorders. In Physician’s Guide to the Diagnosis, Treatment, and Follow-up of Inherited Metabolic Diseases; Blau, N., Duran, M., Gibson, K.M., Dionisi-Vici, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 247–264. ISBN 978-3-642-40337-8. [Google Scholar]
- Grosse, S.D.; Khoury, M.J.; Greene, C.L.; Crider, K.S.; Pollitt, R.J. The epidemiology of medium chain acyl-CoA dehydrogenase deficiency: An update. Genet. Med. 2006, 8, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Rhead, W.J. Newborn screening for medium-chain acyl-CoA dehydrogenase deficiency: A global perspective. J. Inherit. Metab. Dis. 2006, 29, 370–377. [Google Scholar] [CrossRef]
- Onkenhout, W.; Venizelos, V.; van der Poel, P.F.; van den Heuvel, M.P.; Poorthuis, B.J. Identification and quantification of intermediates of unsaturated fatty acid metabolism in plasma of patients with fatty acid oxidation disorders. Clin. Chem. 1995, 41, 1467–1474. [Google Scholar] [CrossRef]
- Scaini, G.; Simon, K.R.; Tonin, A.M.; Busanello, E.N.B.; Moura, A.P.; Ferreira, G.C.; Wajner, M.; Streck, E.L.; Schuck, P.F. Toxicity of octanoate and decanoate in rat peripheral tissues: Evidence of bioenergetic dysfunction and oxidative damage induction in liver and skeletal muscle. Mol. Cell. Biochem. 2012, 361, 329–335. [Google Scholar] [CrossRef]
- Merritt, J.L.; Chang, I.J. Medium-Chain Acyl-Coenzyme a Dehydrogenase Deficiency; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1424/ (accessed on 21 September 2022).
- Ikeda, Y.; Hale, D.E.; Keese, S.M.; Coates, P.M.; Tanaka, K. Biosynthesis of Variant Medium Chain Acyl-CoA Dehydrogenase in Cultured Fibroblasts from Patients with Medium Chain Acyl-CoA Dehydrogenase Deficiency. Pediatr. Res. 1986, 20, 843–847. [Google Scholar] [CrossRef]
- Tajima, G.; Hara, K.; Tsumura, M.; Kagawa, R.; Okada, S.; Sakura, N.; Hata, I.; Shigematsu, Y.; Kobayashi, M. Screening of MCAD deficiency in Japan: 16years’ experience of enzymatic and genetic evaluation. Mol. Genet. Metab. 2016, 119, 322–328. [Google Scholar] [CrossRef]
- Tolwani, R.J.; Hamm, D.A.; Tian, L.; Sharer, J.D.; Vockley, J.; Rinaldo, P.; Matern, D.; Schoeb, T.R.; Wood, P.A. Medium-Chain Acyl-CoA Dehydrogenase Deficiency in Gene-Targeted Mice. PLoS Genet. 2005, 1, e23. [Google Scholar] [CrossRef] [PubMed]
- Platt, S.; McGrotty, Y.L.; Abramson, C.J.; Jakobs, C. Refractory Seizures Associated with an Organic Aciduria in a Dog. J. Am. Anim. Hosp. Assoc. 2007, 43, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, V.; Drögemüller, C.; Leeb, T.; Aguirre, G.; André, C.; Bannasch, D.; Becker, D.; Davis, B.; Ekenstedt, K.; Faller, K.; et al. A comprehensive biomedical variant catalogue based on whole genome sequences of 582 dogs and eight wolves. Anim. Genet. 2019, 50, 695–704. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Teav, T.; Gallart-Ayala, H.; van der Velpen, V.; Mehl, F.; Henry, H.; Ivanisevic, J. Merged Targeted Quantification and Untargeted Profiling for Comprehensive Assessment of Acylcarnitine and Amino Acid Metabolism. Anal. Chem. 2019, 91, 11757–11769. [Google Scholar] [CrossRef]
- Gramer, G.; Haege, G.; Fang-Hoffmann, J.; Hoffmann, G.F.; Bartram, C.R.; Hinderhofer, K.; Burgard, P.; Lindner, M. Medium-Chain Acyl-CoA Dehydrogenase Deficiency: Evaluation of Genotype-Phenotype Correlation in Patients Detected by Newborn Screening. JIMD Rep. 2015, 23, 101–112. [Google Scholar]
- Couce, M.L.; Sánchez-Pintos, P.; Diogo, L.; Leão-Teles, E.; Martins, E.; Santos, H.; Bueno, M.A.; Delgado-Pecellín, C.; Castiñeiras, D.E.; Cocho, J.A.; et al. Newborn screening for medium-chain acyl-CoA dehydrogenase deficiency: Regional experience and high incidence of carnitine deficiency. Orphanet J. Rare Dis. 2013, 8, 1–6. [Google Scholar] [CrossRef]
- Knowler, S.P.; Galea, G.L.; Rusbridge, C. Morphogenesis of Canine Chiari Malformation and Secondary Syringomyelia: Disorders of Cerebrospinal Fluid Circulation. Front. Vet. Sci. 2018, 5, 1–15. [Google Scholar] [CrossRef]
- Rusbridge, C.; Greitz, D.; Iskandar, B.J. Syringomyelia: Current concepts in pathogenesis, diagnosis, and treatment. J. Vet. Intern. Med. 2006, 20, 469–479. [Google Scholar] [CrossRef]
- Driver, C.J.; Chandler, K.; Walmsley, G.; Shihab, N.; Volk, H.A. The association between Chiari-like malformation, ventriculomegaly and seizures in cavalier King Charles spaniels. Vet. J. 2013, 195, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, T.; Monteith, G.; Cortez, M.A.; Wielaender, F.; Fischer, A.; Jokinen, T.S.; Lohi, H.; Sanders, S.; Sammut, V.; Tai, T.; et al. Effect of prior general anesthesia or sedation and antiseizure drugs on the diagnostic utility of wireless video electroencephalography in dogs. J. Vet. Intern. Med. 2020, 34, 1967–1974. [Google Scholar] [CrossRef] [PubMed]
- Wilcken, B.; Haas, M.; Joy, P.; Wiley, V.; Chaplin, M.; Black, C.; Fletcher, J.; McGill, J.; Boneh, A. Outcome of neonatal screening for medium-chain acyl-CoA dehydrogenase deficiency in Australia: A cohort study. Lancet 2007, 369, 37–42. [Google Scholar] [CrossRef]
- Korman, S.H.; Gutman, A.; Brooks, R.; Sinnathamby, T.; Gregersen, N.; Andresen, B.S. Homozygosity for a severe novel medium-chain acyl-CoA dehydrogenase (MCAD) mutation IVS3-1G > C that leads to introduction of a premature termination codon by complete missplicing of the MCAD mRNA and is associated with phenotypic diversity ranging from sudden neonatal death to asymptomatic status. Mol. Genet. Metab. 2004, 82, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Nohara, F.; Tajima, G.; Sasai, H.; Makita, Y. MCAD deficiency caused by compound heterozygous pathogenic variants in ACADM. Hum. Genome Var. 2022, 9, 2. [Google Scholar] [CrossRef]
- Mugnier, A.; Mila, H.; Guiraud, F.; Brévaux, J.; Lecarpentier, M.; Martinez, C.; Mariani, C.; Adib-Lesaux, A.; Chastant-Maillard, S.; Saegerman, C.; et al. Birth weight as a risk factor for neonatal mortality: Breed-specific approach to identify at-risk puppies. Prev. Vet. Med. 2019, 171, 104746. [Google Scholar] [CrossRef]
- Tønnessen, R.; Borge, K.S.; Nødtvedt, A.; Indrebø, A. Canine perinatal mortality: A cohort study of 224 breeds. Theriogenology 2012, 77, 1788–1801. [Google Scholar] [CrossRef]
| Filtering Step | Homozygous Variants |
|---|---|
| All variants in the affected dog | 3,063,158 |
| Private variants | 1562 |
| Protein-changing private variants | 10 |
| Private protein changing variants in ACADM candidate gene | 3 |
| Genotype Frequency | wt/wt | wt/mut | mut/mut |
|---|---|---|---|
| Number (Percentage) of dogs | 98 (60.5%) | 52 (32.1%) | 12 (7.4%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christen, M.; Bongers, J.; Mathis, D.; Jagannathan, V.; Quintana, R.G.; Leeb, T. ACADM Frameshift Variant in Cavalier King Charles Spaniels with Medium-Chain Acyl-CoA Dehydrogenase Deficiency. Genes 2022, 13, 1847. https://doi.org/10.3390/genes13101847
Christen M, Bongers J, Mathis D, Jagannathan V, Quintana RG, Leeb T. ACADM Frameshift Variant in Cavalier King Charles Spaniels with Medium-Chain Acyl-CoA Dehydrogenase Deficiency. Genes. 2022; 13(10):1847. https://doi.org/10.3390/genes13101847
Chicago/Turabian StyleChristen, Matthias, Jos Bongers, Déborah Mathis, Vidhya Jagannathan, Rodrigo Gutierrez Quintana, and Tosso Leeb. 2022. "ACADM Frameshift Variant in Cavalier King Charles Spaniels with Medium-Chain Acyl-CoA Dehydrogenase Deficiency" Genes 13, no. 10: 1847. https://doi.org/10.3390/genes13101847
APA StyleChristen, M., Bongers, J., Mathis, D., Jagannathan, V., Quintana, R. G., & Leeb, T. (2022). ACADM Frameshift Variant in Cavalier King Charles Spaniels with Medium-Chain Acyl-CoA Dehydrogenase Deficiency. Genes, 13(10), 1847. https://doi.org/10.3390/genes13101847






