Comparison on Major Gene Mutations Related to Rifampicin and Isoniazid Resistance between Beijing and Non-Beijing Strains of Mycobacterium tuberculosis: A Systematic Review and Bayesian Meta-Analysis
Abstract
:1. Introduction
2. Methods
2.1. Study Design
2.2. Literature Search Strategy
2.3. Study Selection Criteria
2.4. Data Extraction
2.5. Data Synthesis and Statistical Analysis
3. Results
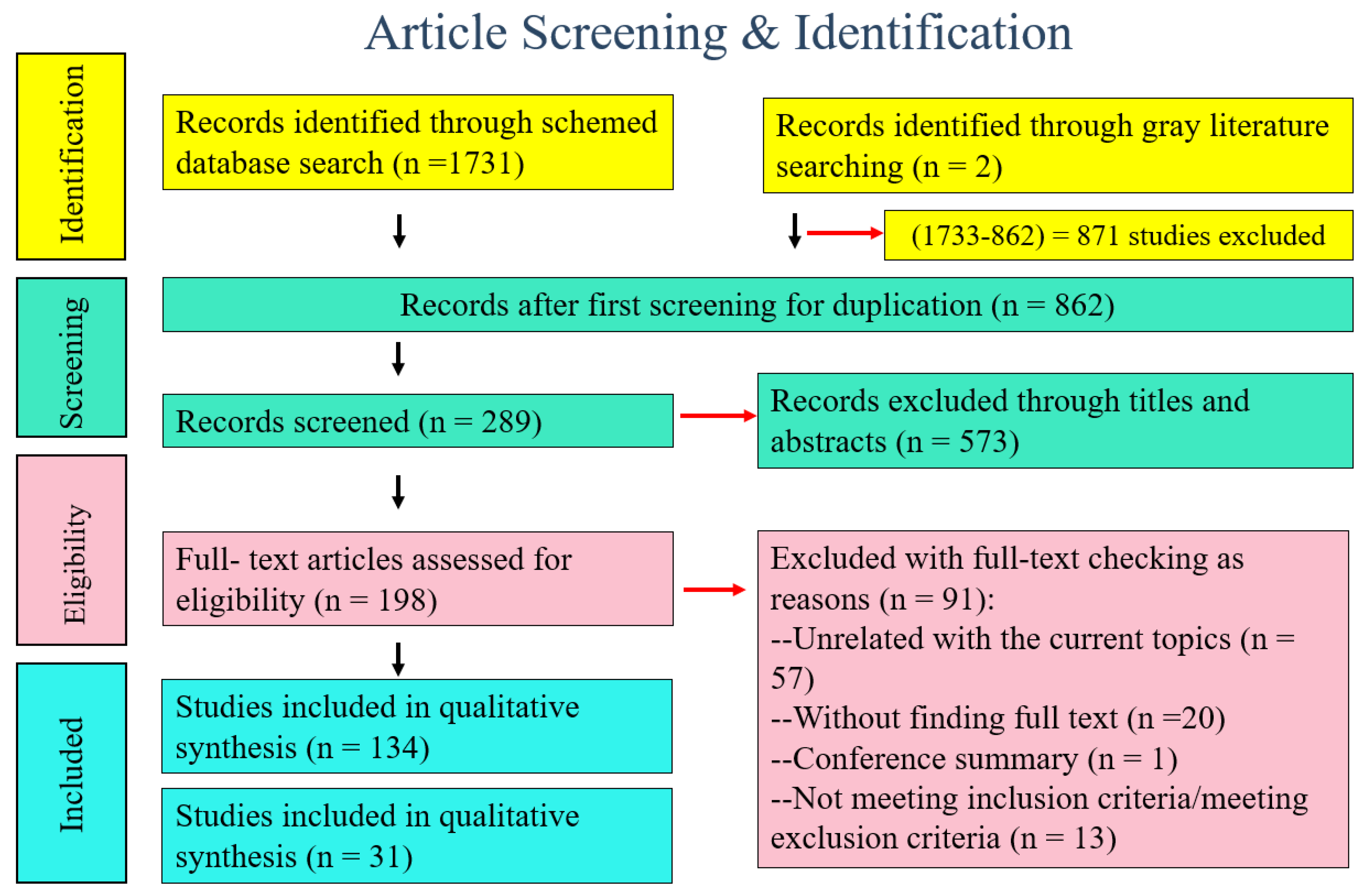
3.1. Literature Search Results
3.2. Characteristics of Studies Included
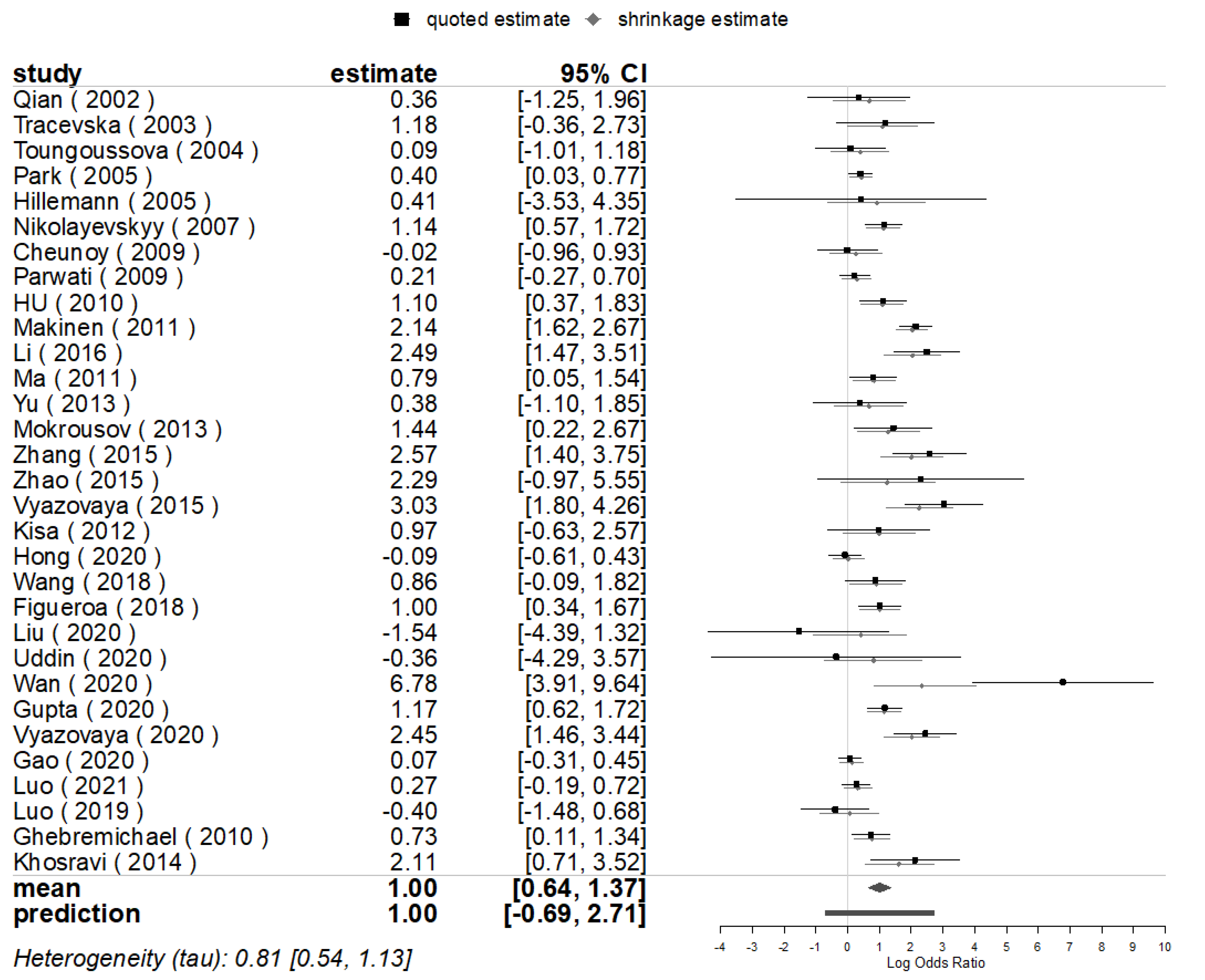
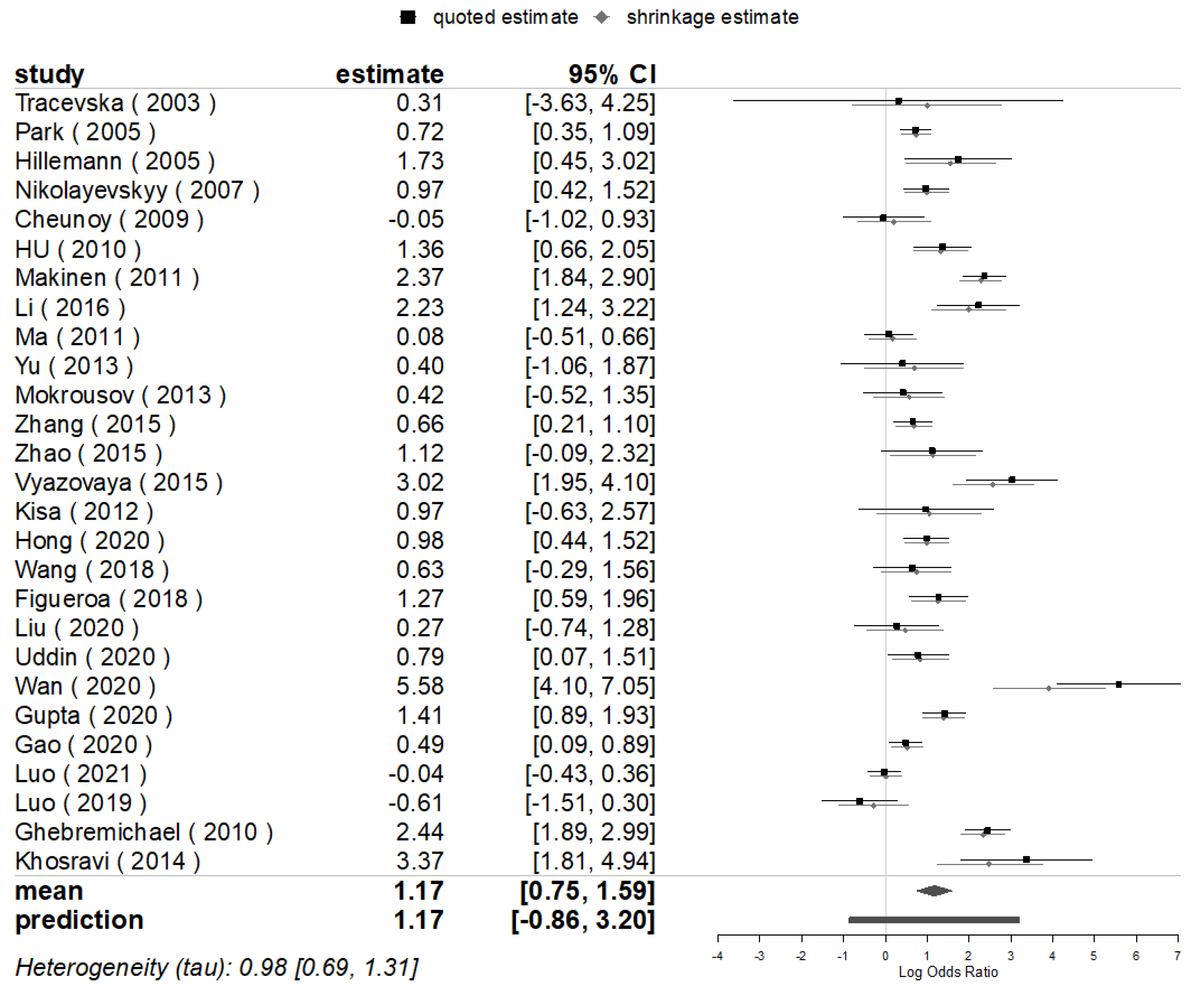
3.3. Mutations Prevalence for Mutations of Genes
3.4. Publication Bias and Sensitivity Analyses
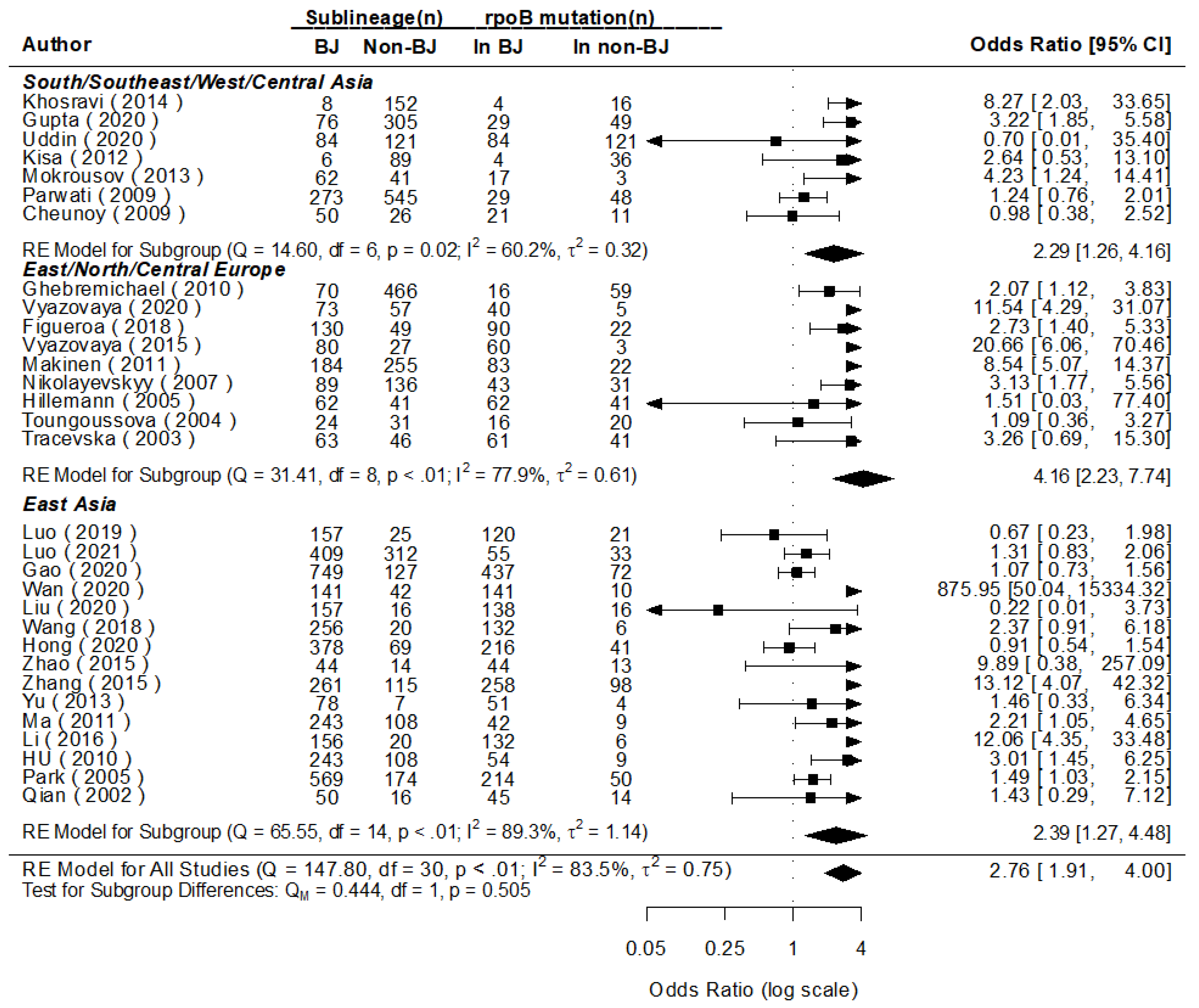
3.5. Mutations of Major Genes in Beijing and Non-Beijing Strains
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alam, A.; Imam, N.; Ahmed, M.M.; Tazyeen, S.; Tamkeen, N.; Farooqui, A.; Malik, M.Z.; Ishrat, R. Identification and Classification of Differentially Expressed Genes and Network Meta-Analysis Reveals Potential Molecular Signatures Associated With Tuberculosis. Front. Genet. 2019, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.M.S.; Tarafder, S.; Anwar, S.; Perdigão, J.; Johora, F.T.; Sattar, H.; Kamal, S.M.M. Circulating strains of Mycobacterium tuberculosis: 24 loci MIRU-VNTR analysis in Bangladesh. Infect. Genet. Evol. 2020, 86, 104634. [Google Scholar] [CrossRef] [PubMed]
- Sann, W.W.M.; Namwat, W.; Faksri, K.; Swe, T.L.; Swe, K.K.; Thwin, T.; Sangka, A. Genetic diversity of Mycobacterium tuberculosis using 24-locus MIRU-VNTR typing and Spoligotyping in Upper Myanmar. J. Infect. Dev. Ctries. 2020, 14, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, X.; Li, W.; Zhang, X.; Wang, W.; Li, C. The study on the association between Beijing genotype family and drug susceptibility phenotypes of Mycobacterium tuberculosis in Beijing. Sci. Rep. 2017, 7, 15076. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Lu, J.; Liu, M.; Wang, Y.; Qu, G.; Li, H.; Wang, J.; Pang, Y.; Liu, C.; Zhao, Y. Genotyping and molecular characteristics of multidrug-resistant Mycobacterium tuberculosis isolates from China. J. Infect. 2015, 70, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Global Tuberculosis Report. 2021. Available online: https://www.who.int/publications-detail-redirect/9789240037021 (accessed on 20 November 2021).
- Parwati, I.; van Crevel, R.; van Soolingen, D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 2010, 10, 103–111. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Musser, J.M. Molecular genetic basis of antimicrobial agent resistance inMycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 1998, 79, 3–29. [Google Scholar] [CrossRef] [Green Version]
- Isakova, J.; Sovkhozova, N.; Vinnikov, D.; Goncharova, Z.; Talaibekova, E.; Aldasheva, N.; Aldashev, A. Mutations of rpoB, katG, inhA and ahp genes in rifampicin and isoniazid-resistant Mycobacterium tuberculosis in Kyrgyz Republic. BMC Microbiol. 2018, 18, 22. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.K.; Shin, S.; Ryu, S.; Cho, S.N.; Koh, W.-J.; Kwon, O.J.; Shim, Y.S.; Lew, W.J.; Bai, G.H. Comparison of drug resistance genotypes between Beijing and non-Beijing family strains of Mycobacterium tuberculosis in Korea. J. Microbiol. Methods 2005, 63, 165–172. [Google Scholar] [CrossRef]
- Ramazanzadeh, R.; Sayhemiri, K. Prevalence of Beijing family in Mycobacterium tuberculosis in world population: Systematic Review and Meta-Analysis. Int. J. Mycobacteriology 2014, 3, 41–45. [Google Scholar] [CrossRef]
- Tarashi, S.; Fateh, A.; Jamnani, F.R.; Siadat, S.D.; Vaziri, F. Prevalence of Beijing and Haarlem genotypes among multidrug-resistant Mycobacterium tuberculosis in Iran: Systematic review and meta-analysis. Tuberculosis 2017, 107, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Garzon-Chavez, D.; Zurita, J.; Mora-Pinargote, C.; Franco-Sotomayor, G.; Leon-Benitez, M.; Granda-Pardo, J.C.; Trueba, G.; Garcia-Bereguiain, M.A.; de Waard, J.H. Prevalence, Drug Resistance, and Genotypic Diversity of the Mycobacterium tuberculosis Beijing Family in Ecuador. Microb. Drug Resist. 2019, 25, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Almeida Da Silva, P.E.; Palomino, J.C. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: Classical and new drugs. J. Antimicrob. Chemother. 2011, 66, 1417–1430. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, Y.; Zhang, X.; Zhang, Z.; Xing, Q.; Ren, W.; Yao, C.; Yu, J.; Ding, B.; Wang, S.; et al. Evaluation of the frequency of mutation genes in multidrug-resistant tuberculosis (MDR-TB) strains in Beijing, China. Epidemiol. Infect. 2021, 149, e21. [Google Scholar] [CrossRef] [PubMed]
- AlMatar, M.; Var, I.; Kayar, B.; Köksal, F. Differential Expression of Resistant and Efflux Pump Genes in MDR-TB Isolates. Endocr. Metab. Immune Disord. Drug Targets 2020, 20, 271–287. [Google Scholar] [CrossRef]
- Gupta, A.; Sinha, P.; Nema, V.; Gupta, P.; Chakraborty, P.; Kulkarni, S.; Rastogi, N.; Anupurba, S. Detection of Beijing strains of MDR M. tuberculosis and their association with drug resistance mutations in katG, rpoB, and embB genes. BMC Infect. Dis. 2020, 20, 752. [Google Scholar] [CrossRef]
- Li, Q.J.; Jiao, W.W.; Yin, Q.Q.; Xu, F.; Li, J.Q.; Sun, L.; Xiao, J.; Li, Y.J.; Mokrousov, I.; Huang, H.R.; et al. Compensatory Mutations of Rifampin Resistance Are Associated with Transmission of Multidrug-Resistant Mycobacterium tuberculosis Beijing Genotype Strains in China. Antimicrob. Agents Chemother. 2016, 60, 2807–2812. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, J.; Shen, J.; Feng, X.; Liu, W.; Zhu, D.; Zheng, H.; Hu, D. Genotyping and Molecular Characterization of Fluoroquinolone’s Resistance Among Multidrug-Resistant Mycobacterium tuberculosis in Southwest of China. Microb. Drug Resist. 2021, 27, 865–870. [Google Scholar] [CrossRef]
- Gao, M.; Yang, T.; Li, G.; Chen, R.; Liu, H.; Gao, Q.; Wan, K.; Feng, S. Analysis on drug resistance-associated mutations of multi-drug resistant Mycobacterium tuberculosis based on whole-genome sequencing in China. Zhong Hua Liuxing Bing Xue Za Zhi 2020, 41, 770–775. [Google Scholar]
- Uddin, M.K.M.; Rahman, A.; Ather, M.F.; Ahmed, T.; Rahman, S.M.M.; Ahmed, S.; Banu, S. Distribution and Frequency of rpoB Mutations Detected by Xpert MTB/RIF Assay Among Beijing and Non-Beijing Rifampicin Resistant Mycobacterium tuberculosis Isolates in Bangladesh. IDR 2020, 13, 789–797. [Google Scholar] [CrossRef] [Green Version]
- Ghebremichael, S.; Groenheit, R.; Pennhag, A.; Koivula, T.; Andersson, E.; Bruchfeld, J.; Hoffner, S.; Romanus, V.; Källenius, G. Drug Resistant Mycobacterium tuberculosis of the Beijing Genotype Does Not Spread in Sweden. PLoS ONE 2010, 5, e10893. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A. Genetics of Drug Resistance in Tuberculosis. Clin. Chest Med. 1997, 18, 55–64. [Google Scholar] [CrossRef]
- Yu, X.; Wen, Z.; Chen, G.; Li, R.; Ding, B.; Yao, Y.; Li, Y.; Wu, H.; Guo, X.; Wang, H.; et al. Molecular characterization of multidrug-resistant Mycobacterium tuberculosis isolated from South-central in China. J. Antibiot. 2014, 67, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Doustdar, F.; Khosravi, A.D.; Farnia, P.; Masjedi, M.R.; Velayati, A.A. Molecular Analysis of Isoniazid Resistance in Different Genotypes of Mycobacterium tuberculosis Isolates from Iran. Microb. Drug Resist. 2008, 14, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Dalla Costa, E.R.; Ribeiro, M.O.; Silva, M.S.; Arnold, L.S.; Rostirolla, D.C.; Cafrune, P.I.; Espinoza, R.C.; Palaci, M.; Telles, M.A.; Ritacco, V.; et al. Correlations of mutations in katG, oxyR-ahpC and inhA genes and in vitro susceptibility in Mycobacterium tuberculosisclinical strains segregated by spoligotype families from tuberculosis prevalent countries in South America. BMC Microbiol. 2009, 9, 39. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Luo, T.; Sun, G.; Qiao, K.; Sun, G.; DeRiemer, K.; Mei, J.; Gao, Q. Mycobacterium tuberculosis Beijing Strains Favor Transmission but Not Drug Resistance in China. Clin. Infect. Dis. 2012, 55, 1179–1187. [Google Scholar] [CrossRef] [Green Version]
- Röver, C. Bayesian random-effects meta-analysis using the bayesmeta R package. J. Stat. Soft. 2020, 93, 6. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.; Antes, G.; Atkins, D.; Barbour, V.; Barrowman, N.; Berlin, J.; Clark, J.; et al. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Rev. Esp. Nutr. Hum. Diet. 2014, 18, 172–181. [Google Scholar] [CrossRef] [Green Version]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies that Evaluate Health Care interventions: Explanation and Elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef] [Green Version]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B.; et al. Meta-analysis of Observational Studies in EpidemiologyA Proposal for Reporting. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- D’Agostino, R.B. Tutorials in Biostatistics, Tutorials in Biostatistics: Statistical Modelling of Complex Medical Data; John Wiley & Sons: Hoboken, NJ, USA, 2005; ISBN 978-0-470-02371-6. [Google Scholar]
- Zarchev, M.; Ruijne, R.; Mulder, C.; Kamperman, A. Prevalence of adult sexual abuse in men with mental illness: Bayesian meta-analysis. BJPsych Open 2022, 8, e16. [Google Scholar] [CrossRef] [PubMed]
- Grey Literature: What It Is & How to Find It | SFU Library. Available online: https://www.lib.sfu.ca/help/research-assistance/format-type/grey-literature (accessed on 5 September 2022).
- Lajeunesse, M.J. Facilitating systematic reviews, data extraction and meta-analysis with the metagear package for r. Methods Ecol. Evol. 2016, 7, 323–330. [Google Scholar] [CrossRef]
- Pettit, S.; Cresta, E.; Winkley, K.; Purssell, E.; Armes, J. Glycaemic control in people with type 2 diabetes mellitus during and after cancer treatment: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0176941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, W.-C.; Chien, K.-L.; Lin, Y.-L.; Wu, M.-S.; Lin, J.-T.; Wang, H.-P.; Tu, Y.-K. Adjuvant treatments for resected pancreatic adenocarcinoma: A systematic review and network meta-analysis. Lancet Oncol. 2013, 14, 1095–1103. [Google Scholar] [CrossRef]
- Meta-Analysis in R with {Metafor}. 2021. Available online: https://www.youtube.com/watch?v=IkduL5iRdqo (accessed on 17 February 2022).
- Peters, J.L.; Sutton, A.J.; Jones, D.R.; Abrams, K.R.; Rushton, L. Comparison of Two Methods to Detect Publication Bias in Meta-Analysis. JAMA 2006, 295, 676–680. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Wang, Z. Comparison of the Power Difference of Egger’s Test and Begg’s Test and the Reason Analysis. J. Huazhong Univ. Sci. Technol. Med. Ed. 2009, 1, 91–94. [Google Scholar]
- Li, L.; Huang, T.; Wang, Y.; Wang, Z.; Liang, Y.; Huang, T.; Zhang, H.; Sun, W.; Wang, Y. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J. Med. Virol. 2020, 92, 577–583. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. A Nonparametric “Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 2000, 95, 89–98. [Google Scholar] [CrossRef]
- Qian, L.; Abe, C.; Lin, T.-P.; Yu, M.-C.; Cho, S.-N.; Wang, S.; Douglas, J.T. rpoB Genotypes of Mycobacterium tuberculosis Beijing Family Isolates from East Asian Countries. J. Clin. Microbiol. 2002, 40, 1091–1094. [Google Scholar] [CrossRef] [Green Version]
- Tracevska, T.; Jansone, I.; Baumanis, V.; Marga, O.; Lillebaek, T. Prevalence of Beijing genotype in Latvian multidrug-resistant Mycobacterium tuberculosis isolates. Int. J. Tuberc. Lung Dis. 2003, 7, 1097–1103. [Google Scholar]
- Toungoussova, O. Impact of drug resistance on fitness of Mycobacterium tuberculosis strains of the W-Beijing genotype. FEMS Immunol. Med. Microbiol. 2004, 42, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Hillemann, D.; Kubica, T.; Rüsch-Gerdes, S.; Niemann, S. Disequilibrium in Distribution of Resistance Mutations among Mycobacterium tuberculosis Beijing and Non-Beijing Strains Isolated from Patients in Germany. Am. Soc. Microbiol. Antimicrob. Agents Chemother. 2005, 49, 1229–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolayevskyy, V.V.; Brown, T.J.; Bazhora, Y.I.; Asmolov, A.A.; Balabanova, Y.M.; Drobniewski, F.A. Molecular epidemiology and prevalence of mutations conferring rifampicin and isoniazid resistance in Mycobacterium tuberculosis strains from the southern Ukraine. Clin. Microbiol. Infect. 2007, 13, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Cheunoy, W.; Haile, M.; Chaiprasert, A.; Prammananan, T.; Cristea-Fernström, M.; Vondracek, M.; Chryssanthou, E.; Hoffner, S.; Petrini, B. Drug resistance and genotypic analysis of Mycobacterium tuberculosis strains from Thai tuberculosis patients. APMIS 2009, 117, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Parwati, I.; Alisjahbana, B.; Apriani, L.; Soetikno, R.; Ottenhoff, T.; Zanden, A.; Meer, J.; Soolingen, D.; Van Crevel, R. Mycobacterium tuberculosis Beijing Genotype Is an Independent Risk Factor for Tuberculosis Treatment Failure in Indonesia. J. Infect. Dis. 2010, 201, 553–557. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Ma, X.; Graviss, E.A.; Wang, W.; Jiang, W.; Xu, B. A major subgroup of Beijing family Mycobacterium tuberculosis is associated with multidrug resistance and increased transmissibility. Epidemiol. Infect. 2011, 139, 130–138. [Google Scholar] [CrossRef]
- Mäkinen, J.; Marjamäki, M.; Haanperä-Heikkinen, M.; Marttila, H.; Endourova, L.B.; Presnova, S.E.; Mathys, V.; Bifani, P.; Ruohonen, R.; Viljanen, M.K.; et al. Extremely high prevalence of multidrug resistant tuberculosis in Murmansk, Russia: A population-based study. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1119–1126. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, Y.; Zhang, C.-L.; Ji, B.-Y.; Zhang, L.-Z.; Shao, Y.-Z.; Jiang, S.-L.; Suzuki, Y.; Nakajima, C.; Fan, C.-L.; et al. Genotypes and Characteristics of Clustering and Drug Susceptibility of Mycobacterium tuberculosis Isolates Collected in Heilongjiang Province, China▿. J. Clin. Microbiol. 2011, 49, 1354–1362. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Hu, Y.; Jiang, W.; Wang, W.; Xu, B. The cluster and drug-resistant characteristics of Beijing genotype Mycobacterium tuberculosis in eastern rural China. Zhonghua Jie He He Hu Xi Za Zhi 2011, 34, 447–450. [Google Scholar]
- Mokrousov, I.; Isakova, J.; Valcheva, V.; Aldashev, A.; Rastogi, N. Molecular snapshot of Mycobacterium tuberculosis population structure and drug-resistance in Kyrgyzstan. Tuberculosis 2013, 93, 501–507. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, Q.; Zeng, C.; Chen, Y.; Zhao, B.; Liu, H.; Xia, Q.; Zhao, X.; Jiao, W.; Li, G.; et al. Molecular characterisation of extensively drug-resistant Mycobacterium tuberculosis isolates in China. Int. J. Antimicrob. Agents 2015, 45, 137–143. [Google Scholar] [CrossRef]
- Vyazovaya, A.; Mokrousov, I.; Solovieva, N.; Mushkin, A.; Manicheva, O.; Vishnevsky, B.; Zhuravlev, V.; Narvskaya, O. Tuberculous Spondylitis in Russia and Prominent Role of Multidrug-Resistant Clone Mycobacterium tuberculosis Beijing B0/W148. Antimicrob. Agents Chemother. 2015, 59, 2349–2357. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Mathema, B.; Zhao, Q.; Zheng, X.; Li, D.; Jiang, W.; Wang, W.; Xu, B. Comparison of the socio-demographic and clinical features of pulmonary TB patients infected with sub-lineages within the W-Beijing and non-Beijing Mycobacterium tuberculosis. Tuberculosis 2016, 97, 18–25. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.; Li, S.; Wang, H.; Wei, J.; Liu, P.; Wang, J.; Zhang, Z.; Gao, H.; Li, M.; et al. Characterization of Mycobacterium tuberculosis isolates from Hebei, China: Genotypes and drug susceptibility phenotypes. BMC Infect Dis 2016, 16, 107. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Li, Y.; Gao, H.; Zhang, Z.; Liu, Y.; Lu, J.; Dai, E. Genotypicdiversity of drug-resistant Mycobacterium tuberculosis isolates from Hebei, China. Int. J. Clin. Exp. Pathol. 2018, 11, 3744–3752. [Google Scholar] [PubMed]
- Figueroa, M.; Ludannyy, R.; Kravtsova, T.; Popov, S.; Vyazovaya, A.; Mokrousov, I. Analysis of Gene Mutations Associated with Mdr among Mycobacterium tuberculosis Strains Isolated In Moscow Region. Russ. J. Infect. Immun. 2019, 8, 562. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Liu, H.; Li, M.; Jiang, Y.; Zhao, X.; Liu, Z.; Wan, K.; Li, G.; Guan, C. Genomic Analysis Identifies Mutations Concerning Drug-Resistance and Beijing Genotype in Multidrug-Resistant Mycobacterium tuberculosis Isolated From China. Front. Microbiol. 2020, 11, 1444. [Google Scholar] [CrossRef] [PubMed]
- Vyazovaya, A.; Proshina, E.; Gerasimova, A.; Avadenii, I.; Solovieva, N.; Zhuravlev, V.; Narvskaya, O.; Mokrousov, I. Increased transmissibility of Russian successful strain Beijing B0/W148 of Mycobacterium tuberculosis: Indirect clues from history and demographics. Tuberculosis 2020, 122, 101937. [Google Scholar] [CrossRef]
- Luo, D.; Qin, H.; Ye, J.; Zhao, J.; Qin, Y.; Lan, R. Analysis on gene mutation characteristics of drug resistant Mycobacterium tuberculosis and its correlation with genotypes in Guangxi. Chin. J. Antituberc. 2021, 43, 596. [Google Scholar] [CrossRef]
- Liu, Z.; Pang, Y.; Chen, S.; Wu, B.; He, H.; Pan, A.; Wang, X. A First Insight into the Genetic Diversity and Drug Susceptibility Pattern of Mycobacterium tuberculosis Complex in Zhejiang, China. BioMed Res. Int. 2016, 2016, 8937539. [Google Scholar] [CrossRef] [Green Version]
- Devi, K.R.; Pradhan, J.; Bhutia, R.; Dadul, P.; Sarkar, A.; Gohain, N.; Narain, K. Molecular diversity of Mycobacterium tuberculosis complex in Sikkim, India and prediction of dominant spoligotypes using artificial intelligence. Sci. Rep. 2021, 11, 7365. [Google Scholar] [CrossRef]
- Mathuria, J.P.; Srivastava, G.N.; Sharma, P.; Mathuria, B.L.; Ojha, S.; Katoch, V.M.; Anupurba, S. Prevalence of Mycobacterium tuberculosis Beijing genotype and its association with drug resistance in North India. J. Infect. Public Health 2017, 10, 409–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y. Characterization on Genotypes, Drug Susceptibility Phenotypes and Gene Mutations Associated with Drug Resistance of Mycobacterium Tuberculosis Clinical Isolates from Hebei. Master’s Degree, Hebei Medical University, Shijiazhuang, China, 2016. [Google Scholar]
- Khosravi, A.D.; Goodarzi, H.; Alavi, S.M.; Akhond, M.R. Application of Deletion- Targeted Multiplex PCR technique for detection of Mycobacterium tuberculosis Beijing strains in samples from tuberculosis patients. Iran J. Microbiol. 2014, 6, 330–334. [Google Scholar] [PubMed]
- Luo, D.; Chen, Q.; Xiong, G.; Peng, Y.; Liu, T.; Chen, X.; Zeng, L.; Chen, K. Prevalence and molecular characterization of multidrug-resistant M. tuberculosis in Jiangxi province, China. Sci. Rep. 2019, 9, 7315. [Google Scholar] [CrossRef] [PubMed]
- Homolka, S.; Köser, C.; Archer, J.; Rüsch-Gerdes, S.; Niemann, S. Single-Nucleotide Polymorphisms in Rv2629 Are Specific for Mycobacterium tuberculosis Genotypes Beijing and Ghana but Not Associated with Rifampin Resistance. J. Clin. Microbiol. 2009, 47, 223–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwaierjiang, A.; Wang, Q.; Liu, H.; Yin, C.; Xu, M.; Li, M.; Liu, M.; Liu, Y.; Zhao, X.; Liu, J.; et al. Prevalence and Molecular Characteristics Based on Whole Genome Sequencing of Mycobacterium tuberculosis Resistant to Four Anti-Tuberculosis Drugs from Southern Xinjiang, China. Infect. Drug Resist. 2021, 14, 3379–3391. [Google Scholar] [CrossRef] [PubMed]
- Bahrm, A.R.; Titov, L.P.; Tasbiti, A.H.; Yari, S.; Graviss, E.A. High-Level Rifampin Resistance Correlates with Multiple Mutations in the rpoB Gene of Pulmonary Tuberculosis Isolates from the Afghanistan Border of Iran. J. Clin. Microbiol. 2009, 47, 2744–2750. [Google Scholar] [CrossRef] [PubMed]


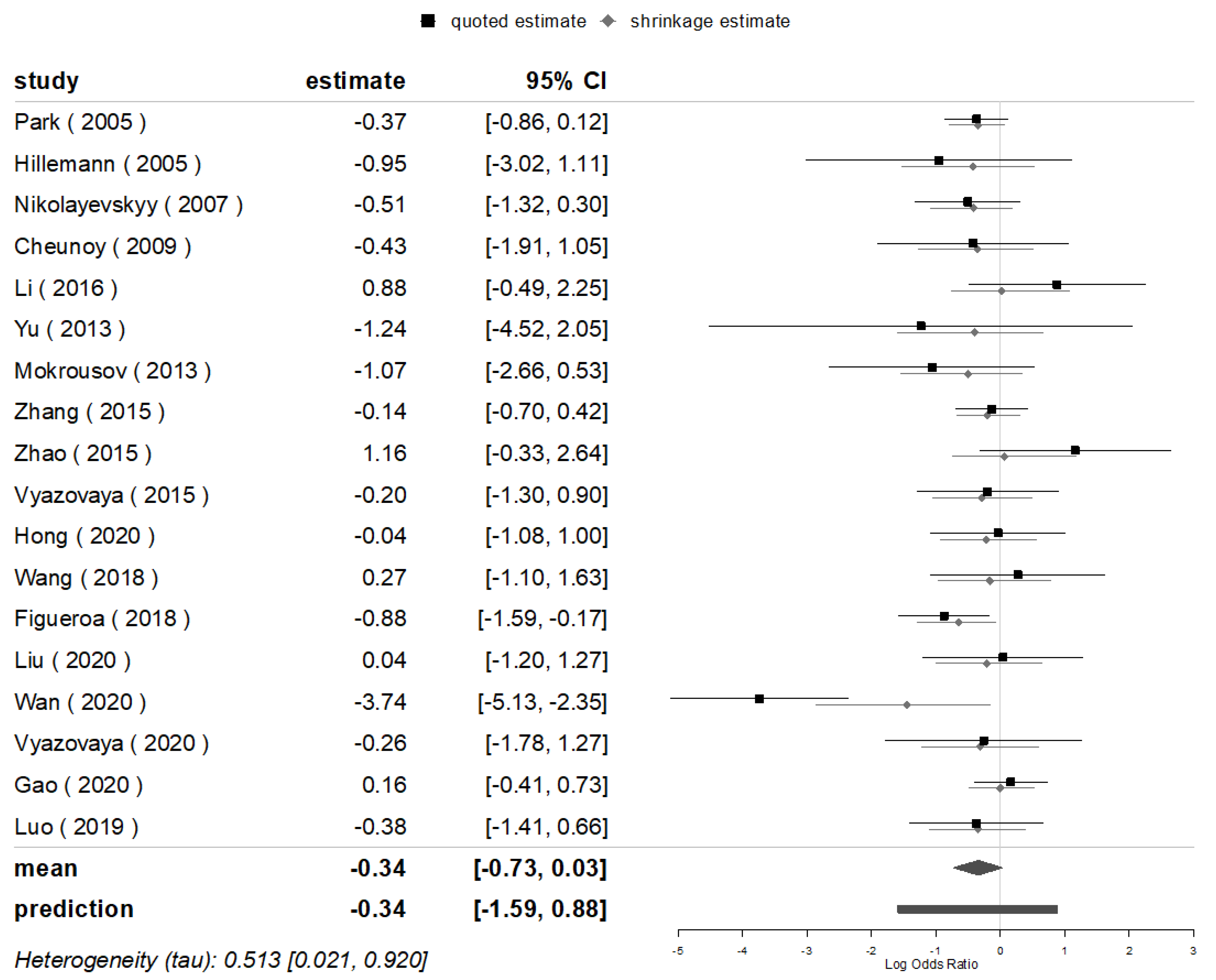
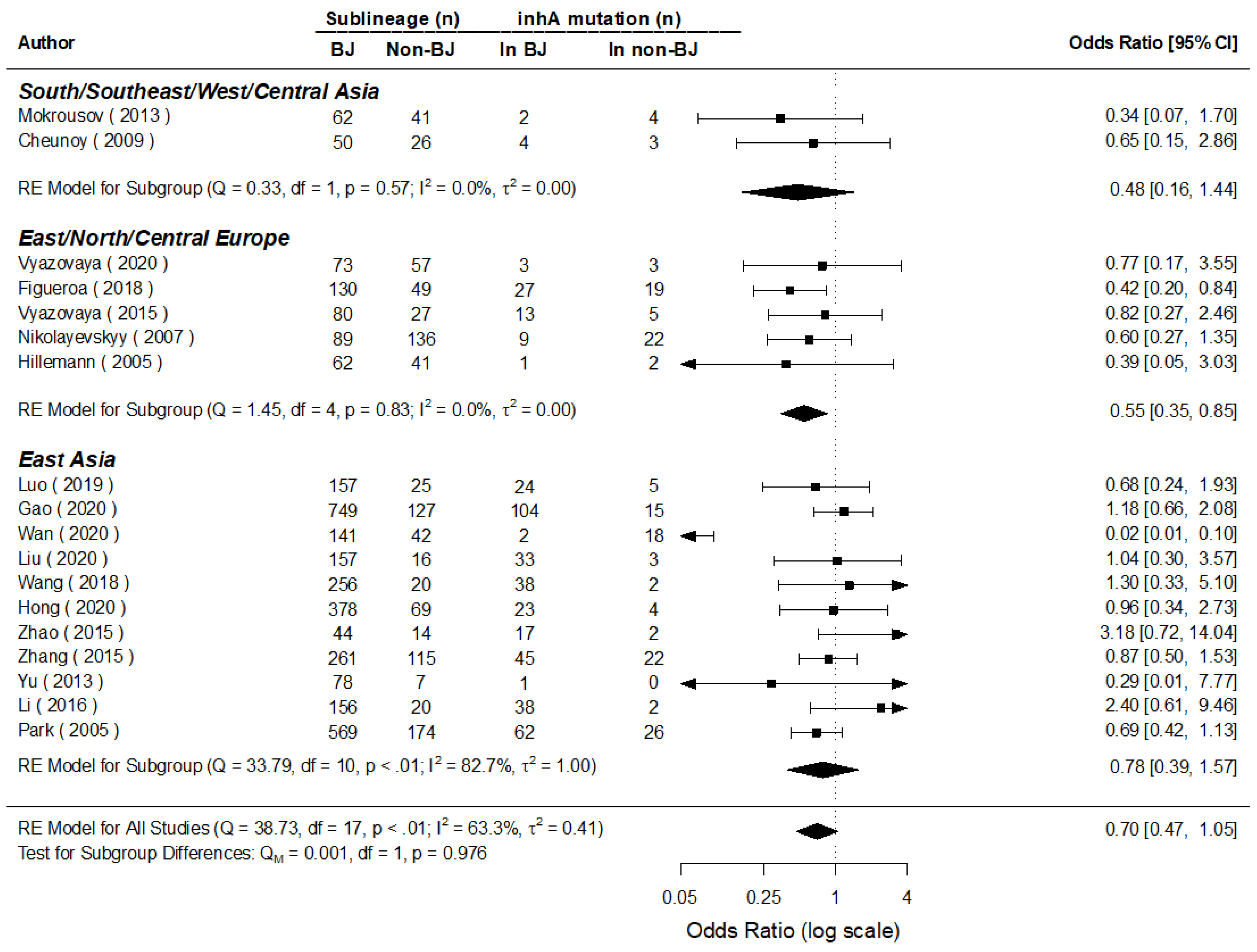
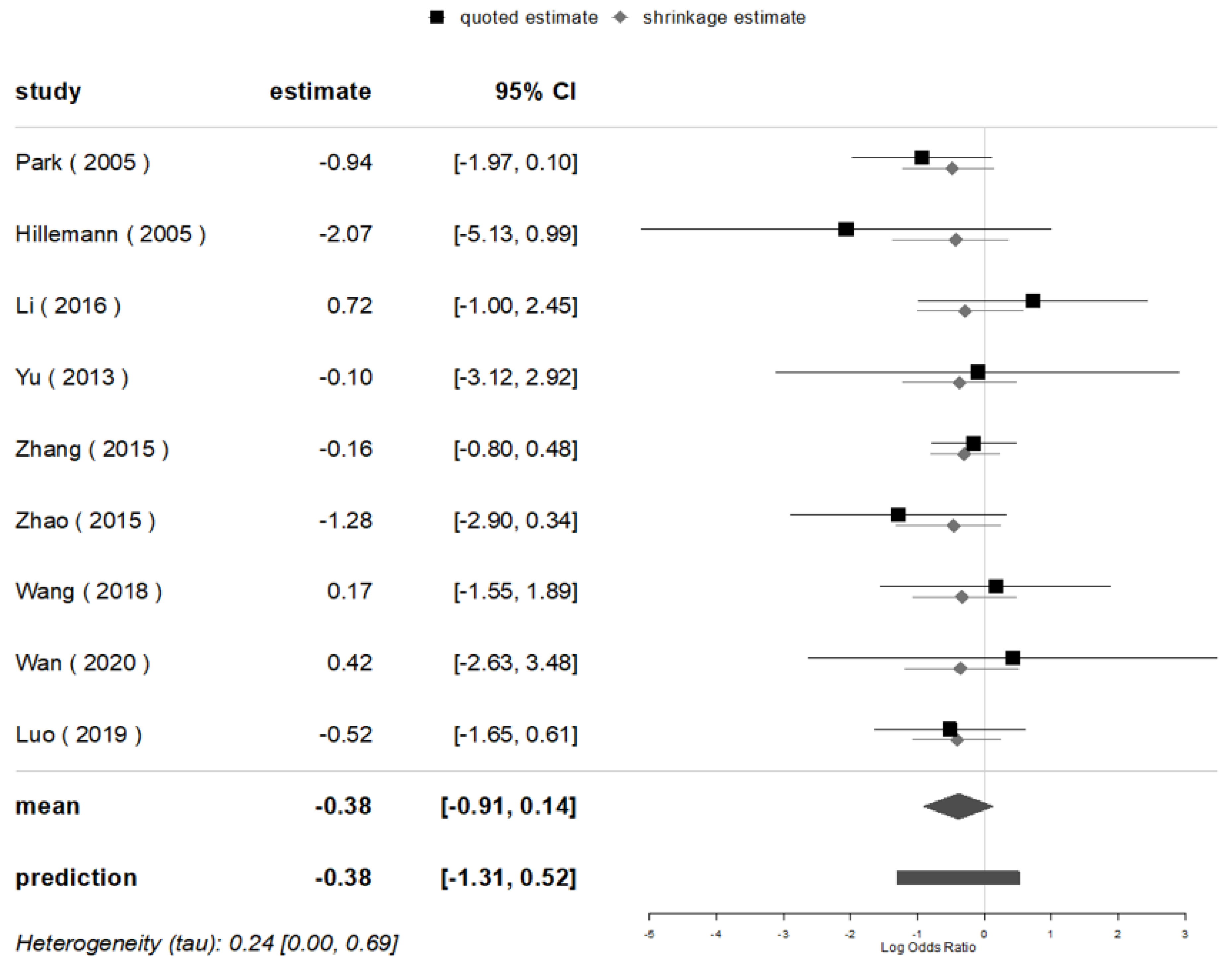

| No. | Author | Country | Year | Isolate Sample Size | Sample Size by Genotypes | rpoB-Rif | katG-INH | inhA-INH | oxyR-ahpC-INH | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | Non-Beijing | Beijing | Non-Beijing | Beijing | Non-Beijing | Beijing | Non-Beijing | Beijing | Non-Beijing | |||||
| 1 | Qian | Asian Countries | 2002 | 66 | 50 | 16 | 45 | 14 | NA | NA | NA | NA | NA | NA |
| 2 | Tracevska | Latvia | 2003 | 109 | 63 | 46 | 61 | 41 | 63 | 46 | NA | NA | NA | NA |
| 3 | Toungoussova | Russia | 2004 | 55 | 24 | 31 | 16 | 20 | NA | NA | NA | NA | NA | NA |
| 4 | Park | Korea | 2005 | 743 | 569 | 174 | 214 | 50 | 250 | 48 | 62 | 26 | 8 | 6 |
| 5 | Hillemann | Germany | 2005 | 103 | 62 | 41 | 62 | 41 | 59 | 31 | 1 | 2 | 0 | 2 |
| 6 | Nikolayevskyy | southern Ukraine | 2007 | 225 | 89 | 136 | 43 | 31 | 52 | 47 | 9 | 22 | NA | NA |
| 7 | Cheunoy | Thailand | 2009 | 76 | 50 | 26 | 21 | 11 | 32 | 17 | 4 | 3 | NA | NA |
| 8 | Parwati | Indonesia | 2009 | 818 | 273 | 545 | 29 | 48 | NA | NA | NA | NA | NA | NA |
| 9 | Hu | China | 2010 | 351 | 243 | 108 | 54 | 9 | 71 | 10 | NA | NA | NA | NA |
| 10 | Mäkinen | Russia | 2011 | 439 | 184 | 255 | 83 | 22 | 91 | 21 | NA | NA | NA | NA |
| 11 | Li | China | 2016 | 176 | 156 | 20 | 132 | 6 | 131 | 7 | 38 | 2 | 21 | 1 |
| 12 | Ma | China | 2011 | 351 | 243 | 108 | 42 | 9 | 46 | 19 | NA | NA | NA | NA |
| 13 | Yu | China | 2013 | 85 | 78 | 7 | 51 | 4 | 42 | 3 | 1 | 0 | 4 | 0 |
| 14 | Mokrousov | Kyrgyzstan | 2013 | 103 | 62 | 41 | 17 | 3 | 17 | 8 | 2 | 4 | NA | NA |
| 15 | Zhang | China | 2015 | 376 | 261 | 115 | 258 | 98 | 173 | 58 | 45 | 22 | 32 | 16 |
| 16 | Zhao | China | 2015 | 58 | 44 | 14 | 44 | 13 | 31 | 6 | 17 | 2 | 3 | 3 |
| 17 | Vyazovaya | Russia | 2015 | 107 | 80 | 27 | 60 | 3 | 71 | 7 | 13 | 5 | NA | NA |
| 18 | Kisa | Turkey | 2012 | 95 | 6 | 89 | 4 | 36 | 4 | 36 | NA | NA | NA | NA |
| 19 | Hong | China | 2020 | 447 | 378 | 69 | 216 | 41 | 301 | 41 | 23 | 4 | NA | NA |
| 20 | Wang | China | 2018 | 276 | 256 | 20 | 132 | 6 | 131 | 7 | 38 | 2 | 21 | 1 |
| 21 | Figueroa | Russia | 2018 | 179 | 130 | 49 | 90 | 22 | 99 | 23 | 27 | 19 | NA | NA |
| 22 | Liu | China | 2020 | 173 | 157 | 16 | 138 | 16 | 98 | 9 | 33 | 3 | NA | NA |
| 23 | Uddin | Bangladesh | 2020 | 205 | 84 | 121 | 84 | 121 | 72 | 88 | NA | NA | NA | NA |
| 24 | Wan | China | 2020 | 183 | 141 | 42 | 141 | 10 | 139 | 7 | 2 | 18 | 2 | 0 |
| 25 | Gupta | India | 2020 | 381 | 76 | 305 | 29 | 49 | 43 | 73 | NA | NA | NA | NA |
| 26 | Vyazovaya | Russia | 2020 | 130 | 73 | 57 | 40 | 5 | NA | NA | 3 | 3 | NA | NA |
| 27 | Gao | China | 2020 | 876 | 749 | 127 | 437 | 72 | 560 | 82 | 104 | 15 | NA | NA |
| 28 | Luo | China | 2021 | 721 | 409 | 312 | 55 | 33 | 65 | 51 | NA | NA | NA | NA |
| 29 | Luo | China | 2019 | 182 | 157 | 25 | 120 | 21 | 90 | 18 | 24 | 5 | 17 | 4 |
| 30 | Ghebremichael | Sweden | 2010 | 536 | 70 | 466 | 16 | 59 | 44 | 59 | 8 | NA | NA | NA |
| 31 | Khosravi | Iran | 2014 | 160 | 8 | 152 | 4 | 16 | 6 | 12 | NA | NA | NA | NA |
| Genes | Sample Size | Mutation Isolates | Mutation Rates | Principal Mutations Pattern | ||||
|---|---|---|---|---|---|---|---|---|
| Total | Beijing | Non-Beijing | Beijing | Non-Beijing | Beijing | Non-Beijing | ||
| rpoB | 8785 | 5225 | 3560 | 2738 | 930 | 52.40 | 26.12 | rpoB Ser531Leu |
| katG | 7716 | 4805 | 2911 | 2781 | 834 | 57.88 | 28.65 | katG S315T |
| inhA | 5034 | 3562 | 1472 | 454 | 157 | 12.75 | 10.67 | inhA -15 C > T, promoter region of inhA |
| oxyR-ahpC | 2182 | 1724 | 458 | 108 | 33 | 6.26 | 7.21 | oxyR-ahpC intergenic region |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Chongsuvivatwong, V.; Lei, S. Comparison on Major Gene Mutations Related to Rifampicin and Isoniazid Resistance between Beijing and Non-Beijing Strains of Mycobacterium tuberculosis: A Systematic Review and Bayesian Meta-Analysis. Genes 2022, 13, 1849. https://doi.org/10.3390/genes13101849
Guo S, Chongsuvivatwong V, Lei S. Comparison on Major Gene Mutations Related to Rifampicin and Isoniazid Resistance between Beijing and Non-Beijing Strains of Mycobacterium tuberculosis: A Systematic Review and Bayesian Meta-Analysis. Genes. 2022; 13(10):1849. https://doi.org/10.3390/genes13101849
Chicago/Turabian StyleGuo, Shengqiong, Virasakdi Chongsuvivatwong, and Shiguang Lei. 2022. "Comparison on Major Gene Mutations Related to Rifampicin and Isoniazid Resistance between Beijing and Non-Beijing Strains of Mycobacterium tuberculosis: A Systematic Review and Bayesian Meta-Analysis" Genes 13, no. 10: 1849. https://doi.org/10.3390/genes13101849
APA StyleGuo, S., Chongsuvivatwong, V., & Lei, S. (2022). Comparison on Major Gene Mutations Related to Rifampicin and Isoniazid Resistance between Beijing and Non-Beijing Strains of Mycobacterium tuberculosis: A Systematic Review and Bayesian Meta-Analysis. Genes, 13(10), 1849. https://doi.org/10.3390/genes13101849





