Multilocus Phylogeography and Population Genetic Analyses of Opsariichthys hainanensis Reveal Pleistocene Isolation Followed by High Gene Flow around the Gulf of Tonkin
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Microsatellite Genotyping, and Mitochondrial Sequencing
2.2. Data Analysis
2.2.1. Mitochondrial DNA and Nuclear DNA Analysis
2.2.2. Microsatellite DNA Analysis
2.2.3. ABC Analyses Using DIYABC
3. Results
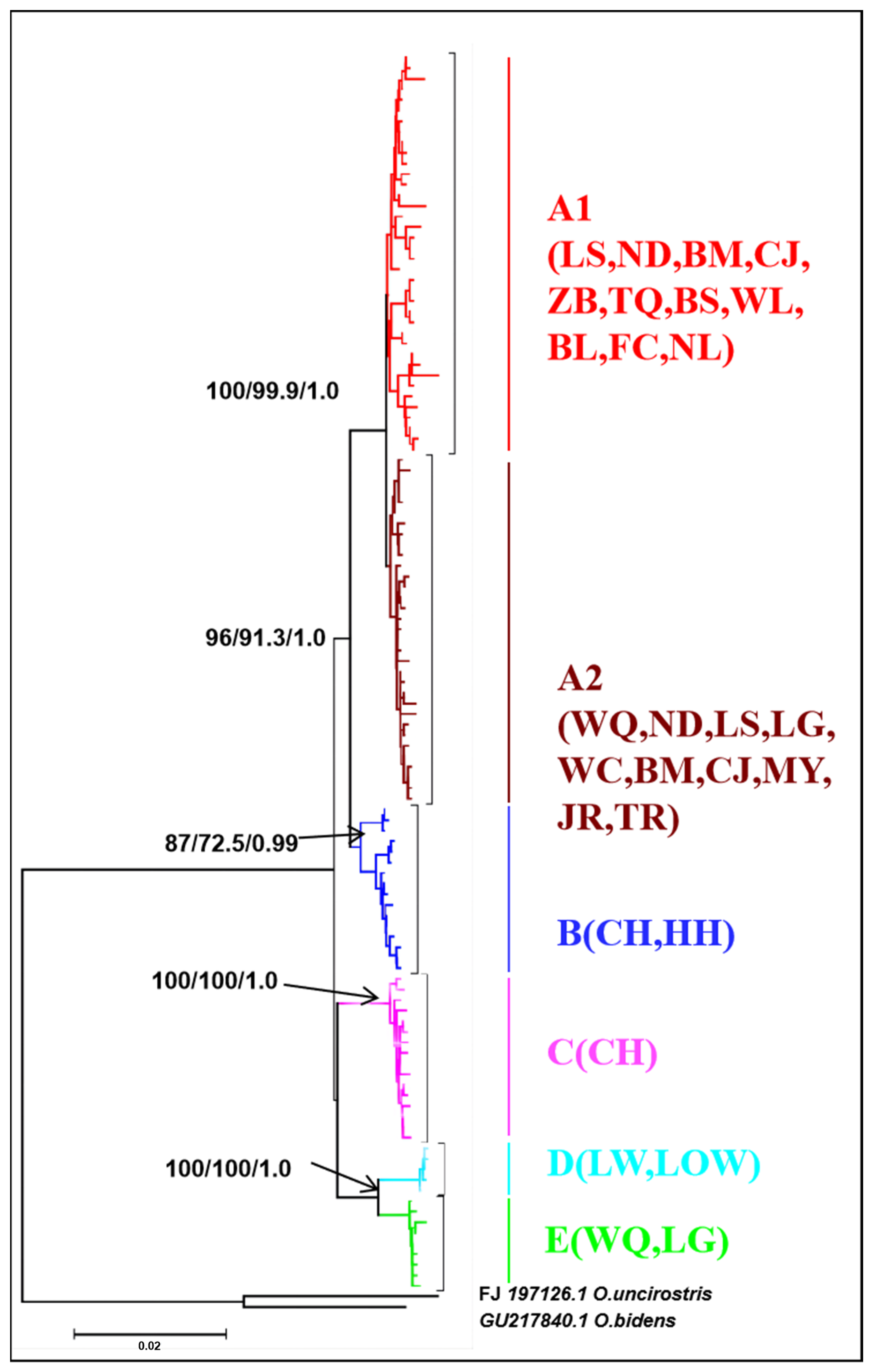
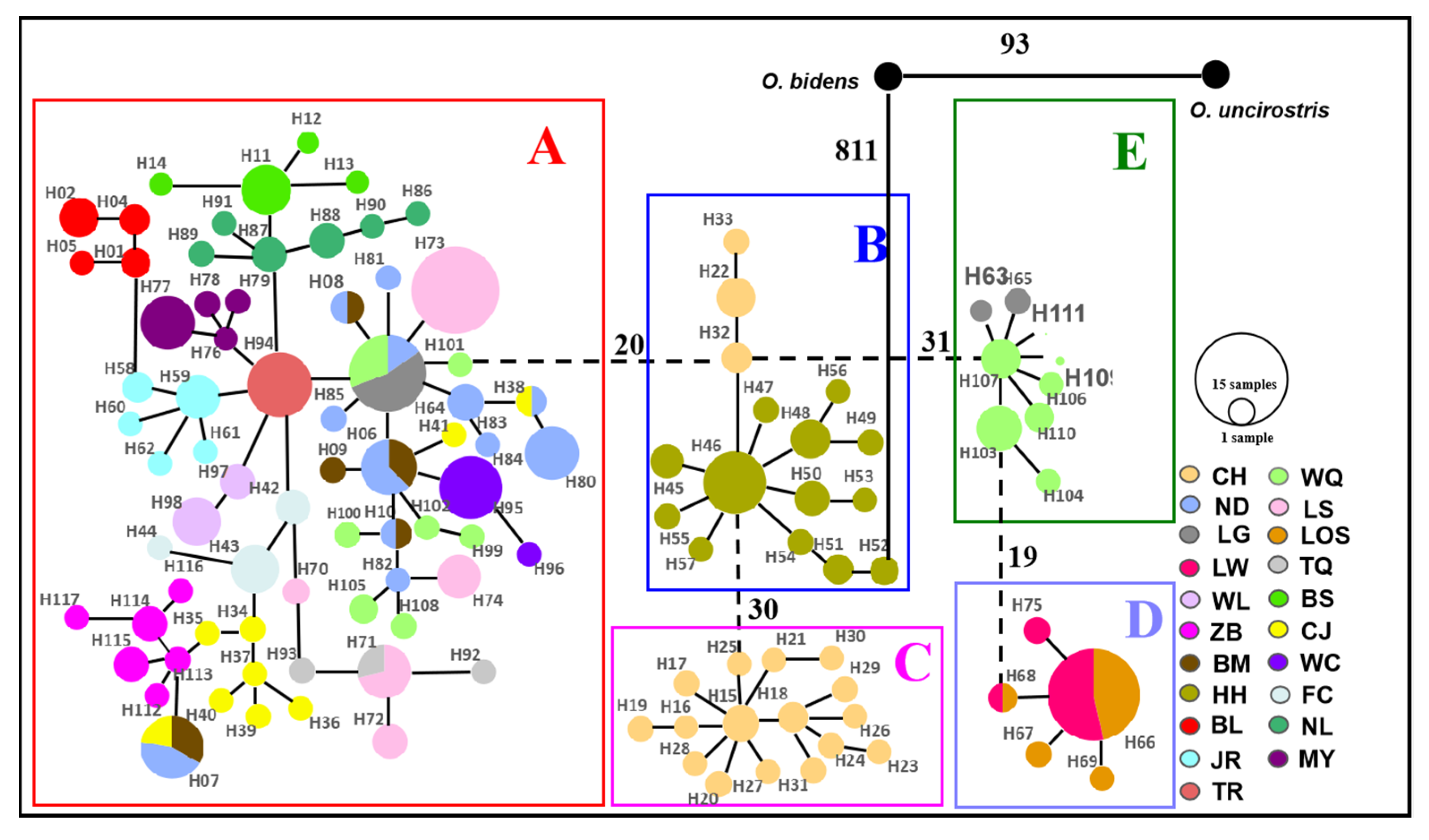
3.1. Mitochondrial DNA and Nuclear DNA Analysis
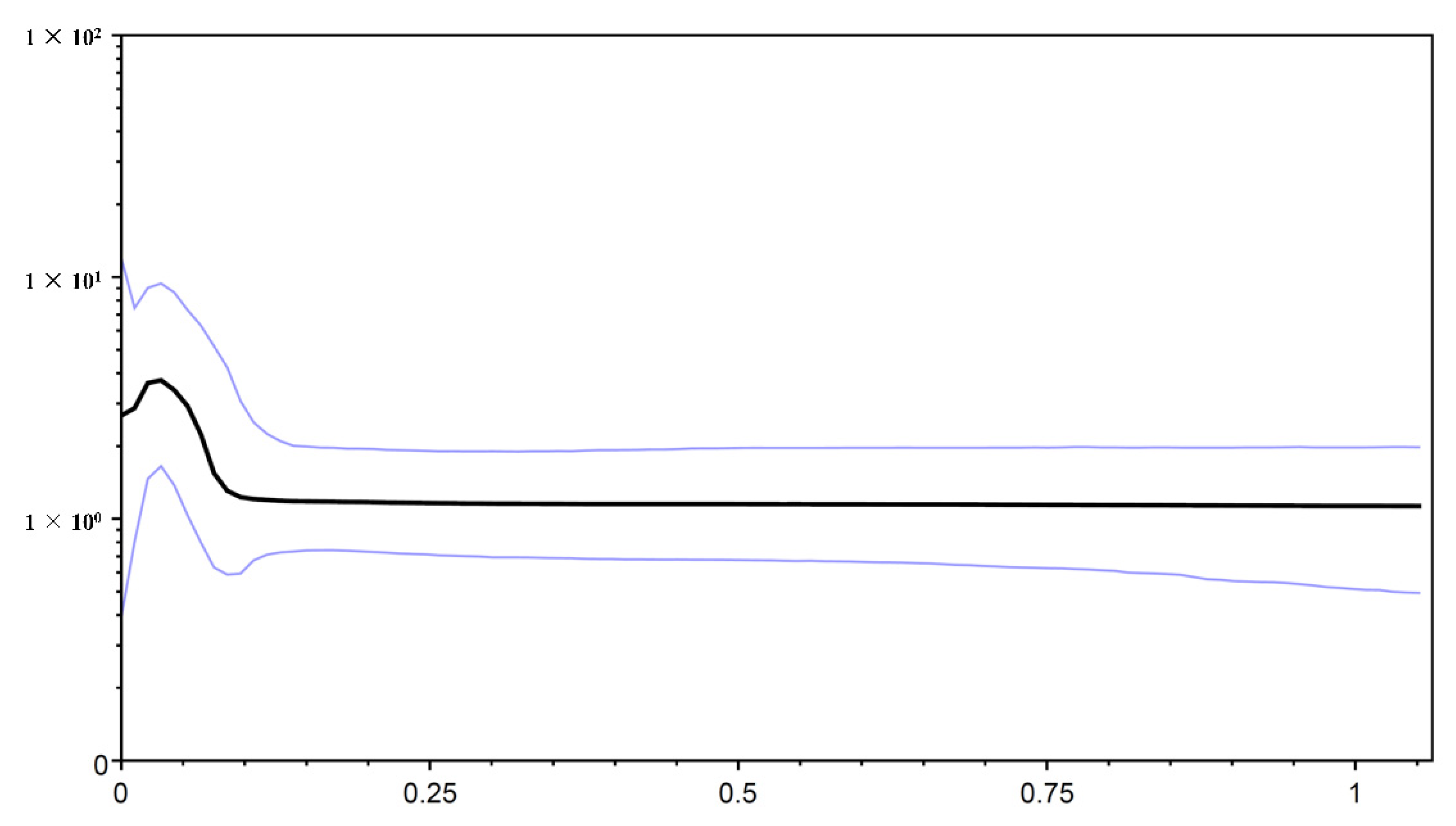
3.2. Historical Population Demography
3.3. Microsatellite DNA
3.4. Approximate Bayesian Computation
4. Discussion
4.1. Genetic Diversity
4.2. Population Structure
4.3. Phylogeography of O. hainanensis
4.4. Demographic History of O. hainanensis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Li, C.; Chen, J.; Wang, J.; Jin, J.; Jiang, S.; Yan, L.; Lin, H.-D.; Zhao, J. Phylogeographic structure of the dwarf snakehead (Channa gachua) around Gulf of Tonkin: Historical biogeography and pronounced effects of sea-level changes. Ecol. Evol. 2021, 11, 12583–12595. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Q.; Tang, W.-Q.; Liao, T.-Y.; Sun, Y.; Zhou, Z.-C.; Han, C.-C.; Liu, D.; Lin, H.-D. Phylogeographical analysis on Squalidus argentatus recapitulates historical landscapes and drainage evolution on the island of Taiwan and mainland China. Int. J. Mol. Sci. 2012, 13, 1405–1425. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-P.; Lin, H.-D.; Huang, S.; Pan, C.-H.; Chen, X.-L.; Chiang, T.-Y. Phylogeography of Varicorhinus barbatulus (Cyprinidae) in Taiwan based on nucleotide variation of mtDNA and allozymes. Mol. Phylogenet. Evol. 2004, 31, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.-Y.; Lin, H.-D.; Zhao, J.; Kuo, P.-H.; Lee, T.-W.; Hsu, K.-C. Diverse processes shape deep phylogeographical divergence in Cobitis sinensis (Teleostei: Cobitidae) in East Asia. J. Zool. Syst. Evol. Res. 2013, 51, 316–326. [Google Scholar] [CrossRef]
- Lin, H.-D.; Kuo, P.-H.; Wang, W.-K.; Chiu, Y.-W.; Ju, Y.-M.; Lin, F.-J.; Hsu, K.-C. Speciation and differentiation of the genus Opsariichthys (Teleostei: Cyprinidae) in East Asia. Biochem. Syst. Ecol. 2016, 68, 92–100. [Google Scholar] [CrossRef]
- Yang, J.-Q.; Hsu, K.-C.; Kuo, P.-H.; Li, L.-L.; Tang, W.-Q.; Liu, D.; Lin, H.-D. Mitochondrial and nuclear genetic structure in Rhodeus ocellatus (Teleostei: Cyprinidae) with approximate Bayesian computation. Environ. Biol. Fishes 2018, 101, 829–841. [Google Scholar] [CrossRef]
- Ju, Y.-M.; Hsu, K.-C.; Yang, J.-Q.; Wu, J.-H.; Li, S.; Wang, W.-K.; Ding, F.; Li, J.; Lin, H.-D. Mitochondrial diversity and phylogeography of Acrossocheilus paradoxus (Teleostei: Cyprinidae). Mitochondrial DNA Part A 2018, 29, 1194–1202. [Google Scholar] [CrossRef]
- Yang, J.-Q.; Hsu, K.-C.; Liu, Z.-Z.; Su, L.-W.; Kuo, P.-H.; Tang, W.-Q.; Zhou, Z.-C.; Liu, D.; Bao, B.-L.; Lin, H.-D. The population history of Garra orientalis (Teleostei: Cyprinidae) using mitochondrial DNA and microsatellite data with approximate Bayesian computation. BMC Evol. Biol. 2016, 16, 73. [Google Scholar] [CrossRef]
- Huang, X.-X.; Hsu, K.-C.; Kang, B.; Kuo, P.-H.; Tsai, W.-H.; Liang, C.-M.; Lin, H.-D.; Wang, W.-K. Population structure of Aphyocypris normalis: Phylogeography and systematics. ZooKeys 2019, 872, 77–90. [Google Scholar] [CrossRef]
- Zhang, W.-J.; Wang, J.-J.; Li, C.; Chen, J.-Q.; Li, W.; Jiang, S.-Y.; Hsu, K.-C.; Zhao, M.-T.; Lin, H.-D.; Zhao, J. Spatial genetic structure of Opsariichthys hainanensis in South China. Mitochondrial DNA Part A 2020, 31, 98–107. [Google Scholar] [CrossRef]
- Economidis, P.S.; Banarescu, P.M. The distribution and origins of freshwater fishes in the Balkan Peninsula, especially in Greece. Int. Rev. Gesamten Hydrobiol. Hydrogr. 1991, 76, 257–284. [Google Scholar] [CrossRef]
- Zhou, T.-Q.; Lin, H.-D.; Hsu, K.-C.; Kuo, P.-H.; Wang, W.-K.; Tang, W.-Q.; Liu, D.; Yang, J.-Q. Spatial genetic structure of the cyprinid fish Onychostoma lepturum on Hainan Island. Mitochondrial DNA Part A 2017, 28, 901–908. [Google Scholar] [CrossRef]
- Voris, H.K. Maps of Pleistocene sea levels in Southeast Asia: Shorelines, river systems and time durations. J. Biogeogr. 2000, 27, 1153–1167. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, Q.; Xie, D.; Fletcher, D.H.; He, D. Factors influencing tropical Island freshwater fishes: Species, status, threats and conservation in Hainan Island. Knowl. Manag. Aquat. Ecosyst. 2018, 419, 6. [Google Scholar] [CrossRef]
- Martinez, A.S.; Willoughby, J.R.; Christie, M.R. Genetic diversity in fishes is influenced by habitat type and life-history variation. Ecol. Evol. 2018, 8, 12022–12031. [Google Scholar] [CrossRef]
- Xiao, W.; Zhang, Y.; Liu, H. Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol. Phylogenetics Evol. 2001, 18, 163–173. [Google Scholar] [CrossRef]
- López, J.A.; Chen, W.-J.; Ortí, G. Esociform phylogeny. Copeia 2004, 2004, 449–464. [Google Scholar] [CrossRef]
- Jeanmougin, F.; Thompson, J.D.; Gouy, M.; Higgins, D.G.; Gibson, T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998, 23, 403–405. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Templeton, A.R. The “Eve” hypotheses: A genetic critique and reanalysis. Am. Anthropol. 1993, 95, 51–72. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambau, A.; Suchard, M. BEAST: 1.8.2. 2013. Available online: http://beast.bio.ed.ac.uk (accessed on 1 June 2022).
- Donaldson, K.A.; Wilson, R.R., Jr. Amphi-panamic geminates of snook (Percoidei: Centropomidae) provide a calibration of the divergence rate in the mitochondrial DNA control region of fishes. Mol. Phylogenetics Evol. 1999, 13, 208–213. [Google Scholar] [CrossRef]
- Zardoya, R.; Doadrio, I. Molecular evidence on the evolutionary and biogeographical patterns of European cyprinids. J. Mol. Evol. 1999, 49, 227–237. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Suchard, M. Tracer v1.6. 2014. Available online: http://beast.bio.ed.ac.uk/tracer (accessed on 29 May 2015).
- Dupanloup, I.; Schneider, S.; Excoffier, L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstr 31772430 uct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenetics Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; ShipLey, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (version 2.9.3): A program to estimate and test gene diversities and fixation indices. 2001. Available online: www.unil.ch/izea/softwares/fstat.html (accessed on 1 June 2022).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Langella, O. Populations 1.2.28: A Population Genetic Software. 1999. Available online: http://www.pge.cnrs-gif.fr/bioinfo/populations/index.php (accessed on 1 June 2022).
- Piry, S.; Luikart, G.; Cornuet, J.-M. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Cornuet, J.-M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.M.; Estoup, A. DIYABC v2. 0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the genetics of populations. In Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978; Volume 4. [Google Scholar]
- Ramos-Onsins, S.E.; Rozas, J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Zhao, J.; Hsu, K.-C.; Luo, J.-Z.; Wang, C.-H.; Chan, B.-P.; Li, J.; Kuo, P.-H.; Lin, H.-D. Genetic diversity and population history of Tanichthys albonubes (Teleostei: Cyprinidae): Implications for conservation. Aquat. Conserv. Mar. Freshw. Ecosyst. 2018, 28, 422–434. [Google Scholar] [CrossRef]
- Jang-Liaw, N.-H.; Tominaga, K.; Zhang, C.; Zhao, Y.; Nakajima, J.; Onikura, N.; Watanabe, K. Phylogeography of the Chinese false gudgeon, Abbottina rivularis, in East Asia, with special reference to the origin and artificial disturbance of Japanese populations. Ichthyol. Res. 2019, 66, 460–478. [Google Scholar] [CrossRef]
- Su, L.-W.; Liu, Z.-Z.; Tang, W.-Q.; Bao, B.-L.; Liu, D.; Yang, J.-Q. Development and characterization of thirteen polymorphic microsatellite markers in the fish Osteochilus salsburyi. Conserv. Genet. Resour. 2014, 6, 33–35. [Google Scholar] [CrossRef]
- Dewoody, J.A.; Avise, J.C. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol. 2000, 56, 461–473. [Google Scholar] [CrossRef]
- Castric, V.; Bernatchez, L.; Belkhir, K.; Bonhomme, F. Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchill (Pisces, Salmonidae): A test of alternative hypotheses. Heredity 2002, 89, 27–35. [Google Scholar] [CrossRef]
- Hurwood, D.A.; Hughes, J.M. Phylogeography of the freshwater fish, Mogurnda adspersa, in streams of northeastern Queensland, Australia: Evidence for altered drainage patterns. Mol. Ecol. 1998, 7, 1507–1517. [Google Scholar] [CrossRef]
- Frankham, R. Do island populations have less genetic variation than mainland populations? Heredity 1997, 78, 311–327. [Google Scholar] [CrossRef]
- Qiu, C.-F.; Lin, Y.-G.; Qin, N.; Zhao, J.; Chen, X.-L. Genetic variation and phylogeography of Micronoemacheilus pulcher populations among basin systems between western South China and Hainan Island. Acta Zool. Sin. 2008, 54, 805–813. [Google Scholar]
- Xiong, W.; Xie, D.; Chen, G.; He, D. Freshwater fish biodiversity in the Leizhou Peninsula of China. Aquat. Ecosyst. Health Manag. 2019, 22, 160–170. [Google Scholar] [CrossRef]
- Zeng, Z.X.; Zeng, X.Z. Physical Geography of Hainan Island. Sci. Beijing 1989, 60–110. (In Chinese) [Google Scholar]
- Zhao, H.-T.; Wang, L.-R.; Yuan, J.-Y. Origin and time of Qiongzhou Strait. Marine Geol. Quat. Geochronol. 2007, 27, 33–40. [Google Scholar]
- Chen, I.-S.; Huang, S.-P.; Jang-Liaw, N.-H.; Shen, C.-N.; Wu, J.-H. Molecular evidence for genetic differentiation of the Opsariichthys bidens complex (Teleostei: Cyprinidae) in Southern China around South China Sea and the validity of Opsariicthys hainanensis. Raffl. Bull Zool. Suppl. 2008, 19, 215–223. [Google Scholar]
- Zuo, Y.L.; Lin, Y.G.; Liang, X.X.; Ma, T.F.; Qing, N. Genetic variation and population differentiation of Liniparhomaloptera disparis (Cypriniformes: Homalopteridae) based on mtDNA sequences in the control region [J]. J. Fish. China 2009, 33, 925–931. [Google Scholar]
- Grant, W.A.S.; Bowen, B.W. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, X.; Liu, Q.; Zhang, H. Genetic variation and Phylogeography of Sinibrama Macrops (Teleostei: Cyprinidae) in Qiantang River Basin, China. Biochem. Syst. Ecol. 2013, 49, 10–20. [Google Scholar] [CrossRef]
- Chen, W.; Zhong, Z.; Dai, W.; Fan, Q.; He, S. Phylogeographic structure, cryptic speciation and demographic history of the sharpbelly (Hemiculter leucisculus), a freshwater habitat generalist from southern China. BMC Evol. Biol. 2017, 17, 216. [Google Scholar] [CrossRef]
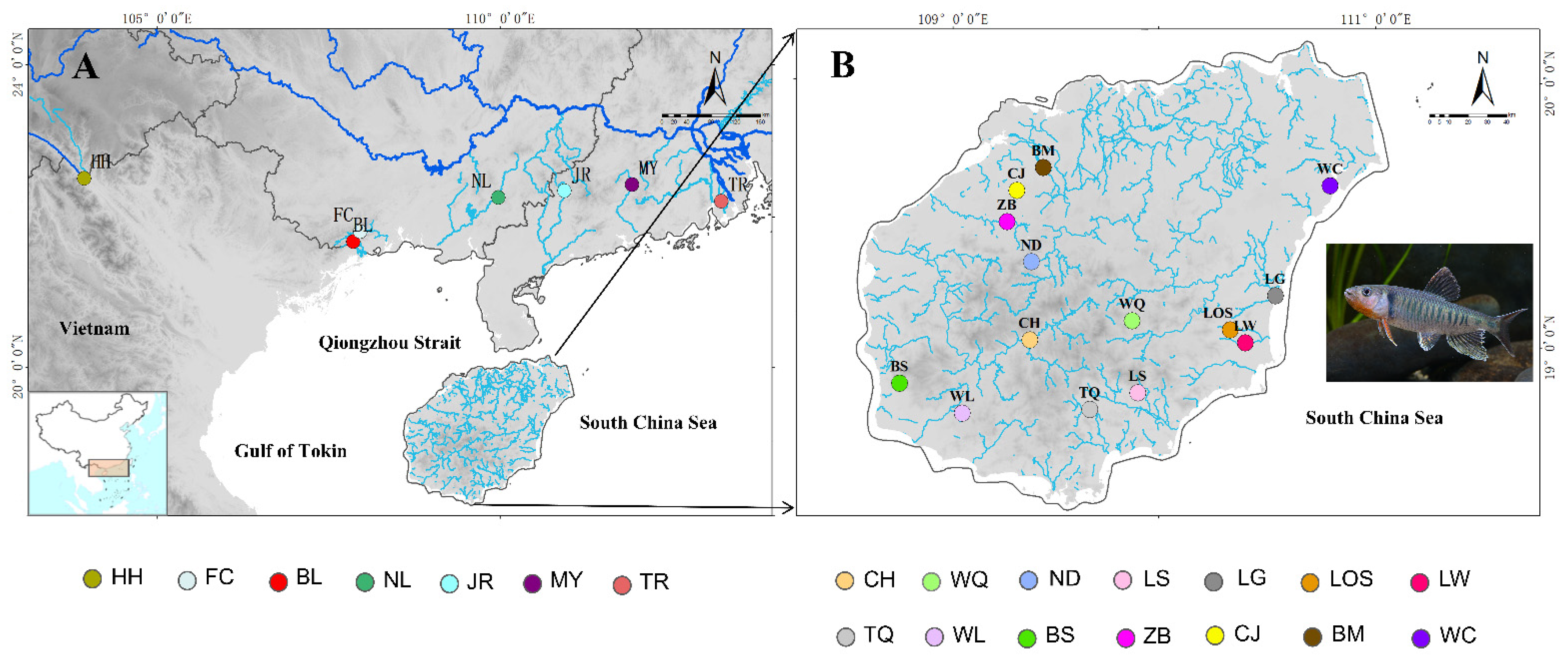


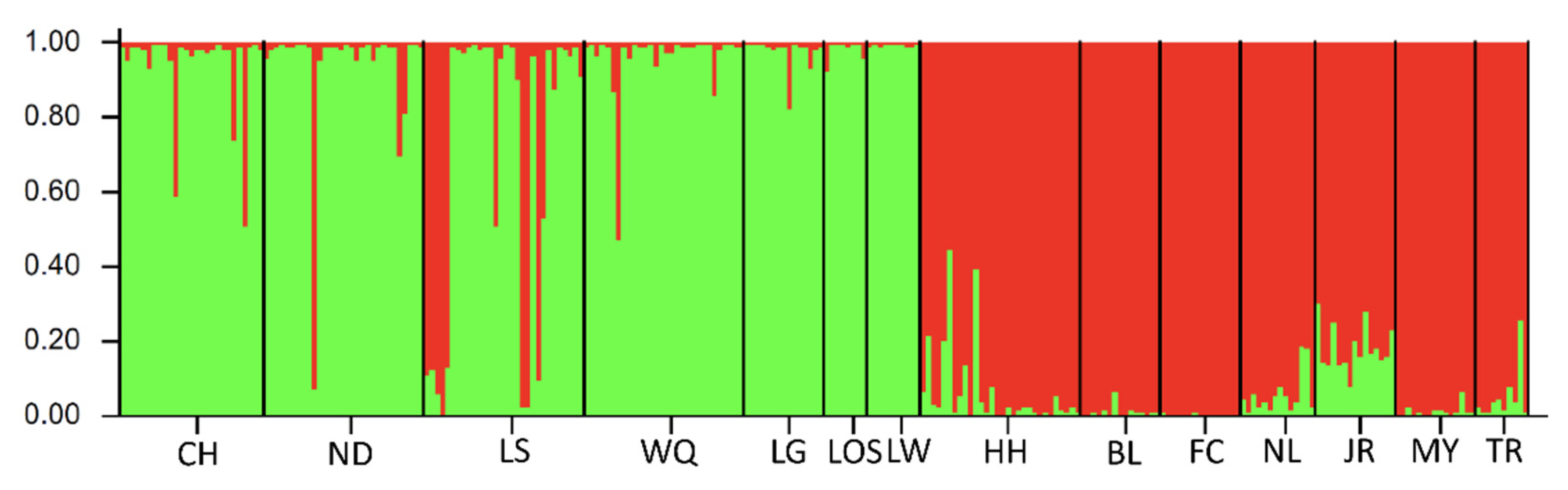
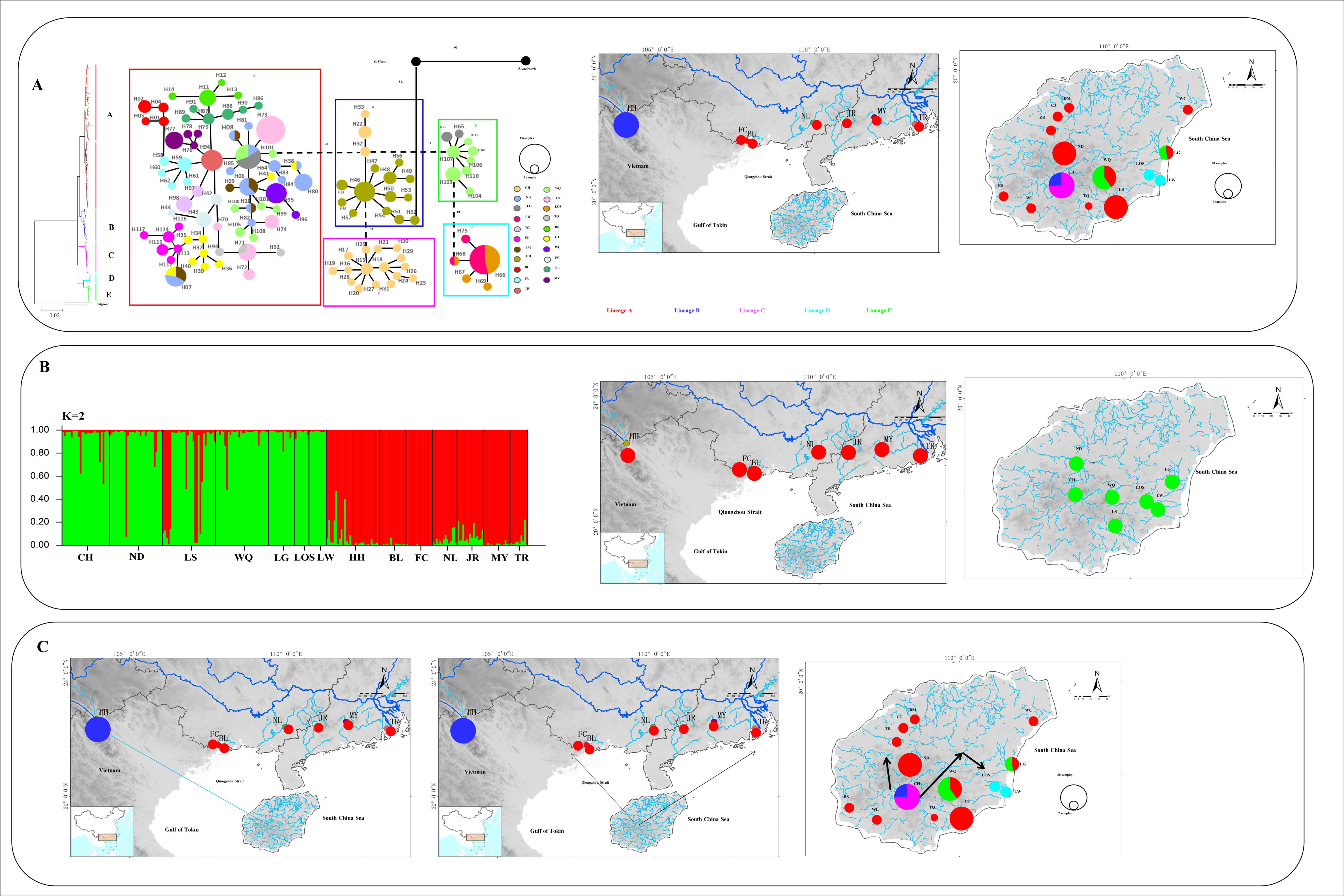
| Gene | Mitochondrial DNA (cyt b Gene and d-Loop Region) | Nuclear DNA (RAG1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locations (Abbreviation) | Sample Size (N) | Haplotype Numbers (Nh) | Haplotype Diversity (h) | Nucleotide Diversity (π) | Nucleotide Diversity (θ) | Sample Size (N) | Haplotype Numbers (Nh) | Haplotype Diversity (h) | Nucleotide Diversity (π) | Nucleotide Diversity (θ) |
| Hainan Island | 215 | 80 | 0.975 | 1.148 | 1.311 | 183 | 45 | 0.908 | 0.199 | 0.850 |
| Changhua River (CH) | 27 | 19 | 0.960 | 0.742 | 0.728 | 24 | 6 | 0.725 | 0.250 | 0.445 |
| Nandu River (ND) | 30 | 12 | 0.897 | 0.204 | 0.244 | 20 | 13 | 0.947 | 0.185 | 0.281 |
| Wanqan River (WQ) | 30 | 14 | 0.924 | 1.062 | 0.635 | 24 | 11 | 0.902 | 0.157 | 0.249 |
| Lingshui River (LS) | 30 | 5 | 0.703 | 0.311 | 0.220 | 20 | 10 | 0.905 | 0.151 | 0.206 |
| Longgun River (LG) | 15 | 3 | 0.600 | 1.071 | 0.640 | 15 | 9 | 0.876 | 0.166 | 0.204 |
| Longshou River (LOS) | 10 | 4 | 0.533 | 0.029 | 0.051 | 9 | 5 | 0.722 | 0.125 | 0.171 |
| Longwei River (LW) | 10 | 3 | 0.378 | 0.019 | 0.034 | 10 | 2 | 0.200 | 0.013 | 0.023 |
| Tengqiao River (TQ) | 4 | 3 | 0.833 | 0.169 | 0.185 | 4 | 4 | 1.000 | 0.243 | 0.254 |
| Wanglou River (WL) | 9 | 2 | 0.500 | 0.024 | 0.018 | 8 | 5 | 0.893 | 0.175 | 0.154 |
| Baisha River (BS) | 10 | 4 | 0.533 | 0.106 | 0.188 | 10 | 3 | 0.378 | 0.027 | 0.047 |
| Zhubi River (ZB) | 10 | 6 | 0.867 | 0.110 | 0.137 | 10 | 3 | 0.378 | 0.134 | 0.164 |
| Chunjiang River (CJ) | 10 | 9 | 0.978 | 0.314 | 0.342 | 9 | 5 | 0.861 | 0.148 | 0.147 |
| Beimen River (BM) | 10 | 5 | 0.844 | 0.267 | 0.222 | 10 | 6 | 0.889 | 0.174 | 0.117 |
| Wenchang River (WC) | 10 | 2 | 0.200 | 0.029 | 0.051 | 10 | 2 | 0.467 | 0.031 | 0.023 |
| Mainland China | 90 | 37 | 0.960 | 0.737 | 0.754 | 74 | 19 | 0.817 | 0.172 | 0.463 |
| Red River (HH) | 30 | 13 | 0.885 | 0.184 | 0.244 | 21 | 2 | 0.095 | 0.006 | 0.018 |
| Beilun River (BL) | 10 | 5 | 0.822 | 0.090 | 0.086 | 10 | 5 | 0.822 | 0.099 | 0.094 |
| Fangheng River (FC) | 10 | 3 | 0.600 | 0.158 | 0.188 | 10 | 3 | 0.378 | 0.053 | 0.094 |
| Nanliu River (NL) | 10 | 6 | 0.867 | 0.125 | 0.154 | 10 | 7 | 0.911 | 0.274 | 0.399 |
| Jian River (JR) | 10 | 5 | 0.756 | 0.075 | 0.120 | 5 | 3 | 0.700 | 0.186 | 0.223 |
| Moyang River (MY) | 10 | 4 | 0.533 | 0.065 | 0.068 | 10 | 4 | 0.533 | 0.103 | 0.164 |
| Tan River (TR) | 10 | 1 | 0.000 | 0.000 | 0.000 | 8 | 4 | 0.821 | 0.164 | 0.128 |
| Total | 305 | 117 | 0.984 | 1.076 | 1.576 | 257 | 59 | 0.897 | 0.194 | 1.073 |
| Mitochondrial cyt b + D-Loop Gene | nuDNA RAG1 Genes | Microsatellite DNA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Scenario Ⅰ: two independent groups divided by the Qiongzhou Strait | |||||||||
| Among groups | 1.24 | FCT = 0.012 | 0.292 | −0.11 | FCT = −0.001 | 0.317 | 1.72 | FCT = 0.017 | 0.004 |
| Among populations within groups | 68.09 | FSC = 0.690 | 0.000 | 30.53 | FSC = 0.305 | 0.000 | 18.24 | FSC = 0.186 | 0.000 |
| Within populations | 30.66 | FST = 0.693 | 0.000 | 69.58 | FST = 0.304 | 0.000 | 80.03 | FST = 0.200 | 0.000 |
| Scenario Ⅱ: three independent groups divided by the Qiongzhou Strait and Beibu Gulf; | |||||||||
| Among groups | 17.14 | FCT = 0.171 | 0.242 | 3.46 | FCT = 0.035 | 0.389 | 3.09 | FCT = 0.031 | 0.370 |
| Among populations within groups | 54.43 | FSC = 0.657 | 0.000 | 26.98 | FSC = 0.279 | 0.000 | 12.31 | FSC = 0.127 | 0.000 |
| Within populations | 28.43 | FST = 0.716 | 0.000 | 69.56 | FST = 0.304 | 0.000 | 84.6 | FST =0.154 | 0.000 |
| Scenario Ⅲ: four groups divided by the WY Range, the Qiongzhou Strait and Beibu Gulf. | |||||||||
| Among groups | 0.35 | FCT = 0.004 | 0.317 | 0.93 | FCT = 0.009 | 0.266 | −1.95 | FCT = −0.019 | 0.635 |
| Among populations within groups | 63.59 | FSC = 0.638 | 0.000 | 24.82 | FSC = 0.251 | 0.000 | 13.99 | FSC = 0.137 | 0.000 |
| Within populations | 36.06 | FST = 0.639 | 0.000 | 74.25 | FST =0.258 | 0.000 | 87.96 | FST = 0.120 | 0.000 |
| SAMOVA | |||||||||
| Among groups | 63.12 | FCT = 0.631 | 0.000 | 31.2 | FCT = 0.312 | 0.000 | 14.93 | FCT = 0.149 | 0.000 |
| Among populations within groups | 7.59 | FSC = 0.206 | 0.000 | 11.45 | FSC = 0.166 | 0.000 | 10.04 | FSC = 0.118 | 0.000 |
| Within populations | 29.28 | FST = 0.707 | 0.000 | 57.35 | FST = 0.426 | 0.000 | 75.03 | FST = 0.250 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zhang, W.; Wu, J.; Li, C.; Ju, Y.-M.; Lin, H.-D.; Zhao, J. Multilocus Phylogeography and Population Genetic Analyses of Opsariichthys hainanensis Reveal Pleistocene Isolation Followed by High Gene Flow around the Gulf of Tonkin. Genes 2022, 13, 1908. https://doi.org/10.3390/genes13101908
Wang J, Zhang W, Wu J, Li C, Ju Y-M, Lin H-D, Zhao J. Multilocus Phylogeography and Population Genetic Analyses of Opsariichthys hainanensis Reveal Pleistocene Isolation Followed by High Gene Flow around the Gulf of Tonkin. Genes. 2022; 13(10):1908. https://doi.org/10.3390/genes13101908
Chicago/Turabian StyleWang, Junjie, Wenjun Zhang, Jinxian Wu, Chao Li, Yu-Min Ju, Hung-Du Lin, and Jun Zhao. 2022. "Multilocus Phylogeography and Population Genetic Analyses of Opsariichthys hainanensis Reveal Pleistocene Isolation Followed by High Gene Flow around the Gulf of Tonkin" Genes 13, no. 10: 1908. https://doi.org/10.3390/genes13101908
APA StyleWang, J., Zhang, W., Wu, J., Li, C., Ju, Y.-M., Lin, H.-D., & Zhao, J. (2022). Multilocus Phylogeography and Population Genetic Analyses of Opsariichthys hainanensis Reveal Pleistocene Isolation Followed by High Gene Flow around the Gulf of Tonkin. Genes, 13(10), 1908. https://doi.org/10.3390/genes13101908







