Comparative Analyses of Plastomes of Four Anubias (Araceae) Taxa, Tropical Aquatic Plants Endemic to Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Plastome Sequencing, Assembly and Annotation
2.2. Comparative Plastome Analysis
2.3. Repeats, Nucletide Diversity, Codon Usage and RNA Editing Sites
2.4. Phylogenetic Analysis
2.5. Positive Selection Analysis
3. Results
3.1. Plastome Features
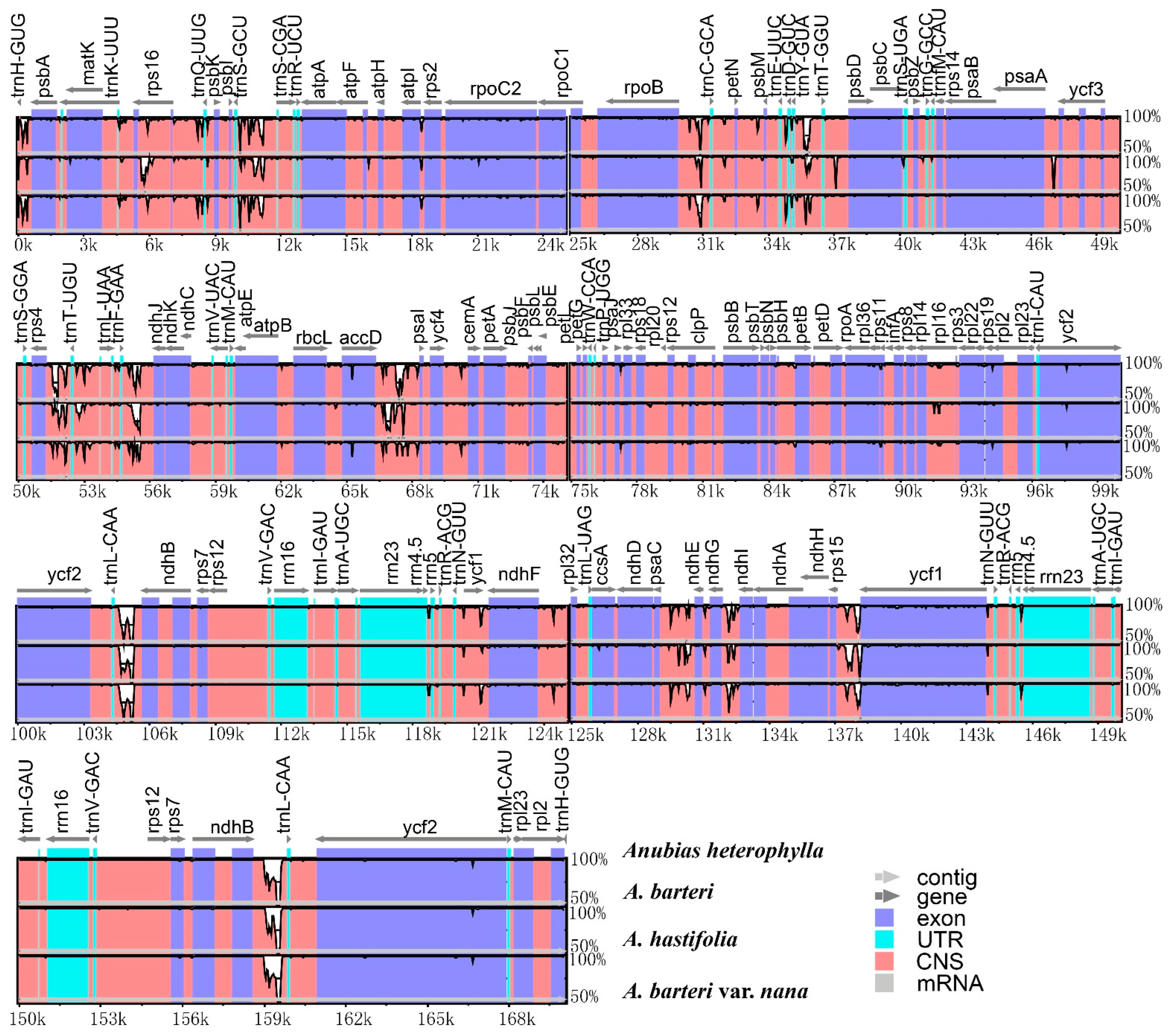
3.2. Contraction and Expansion of INVERTED repeats
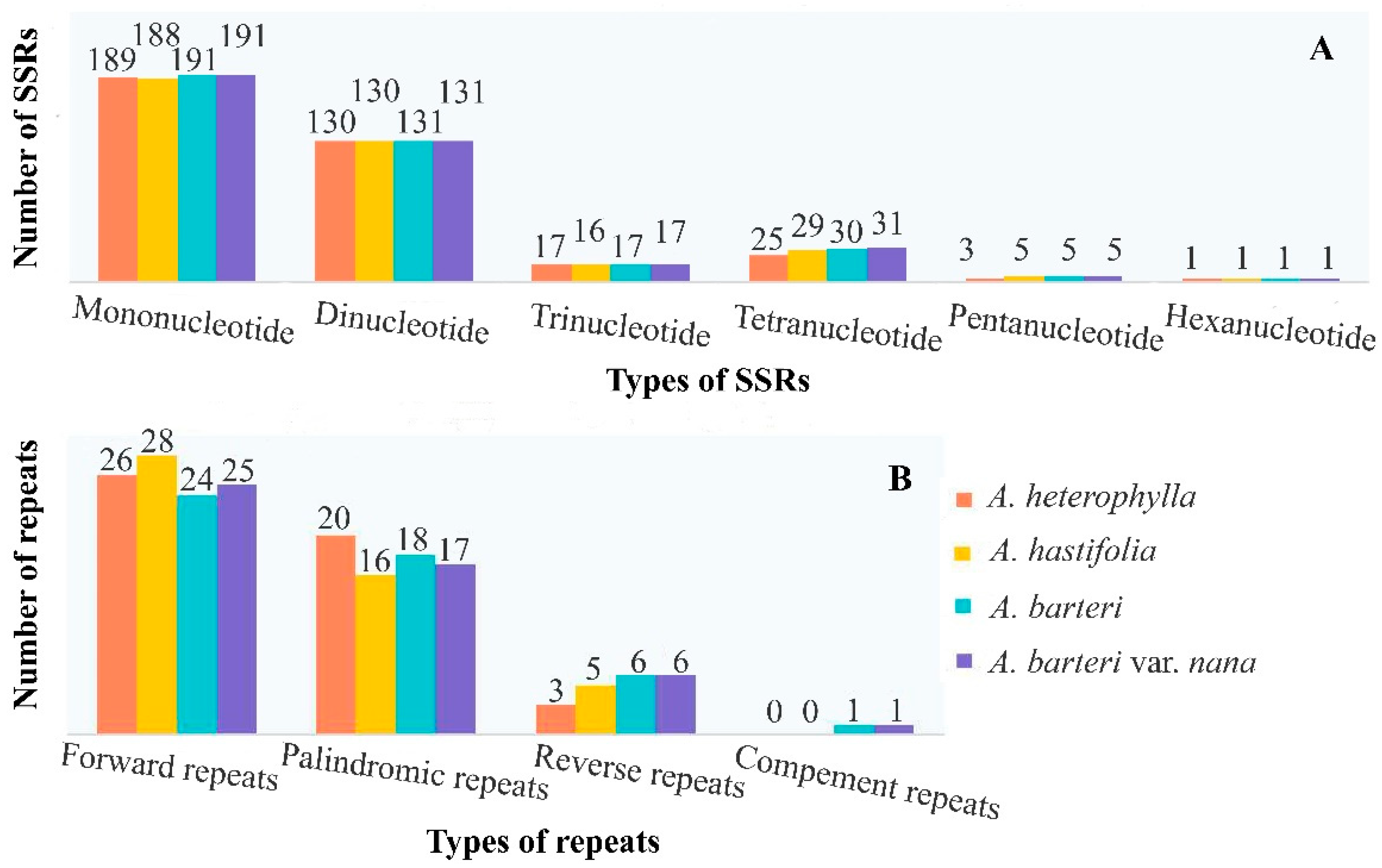
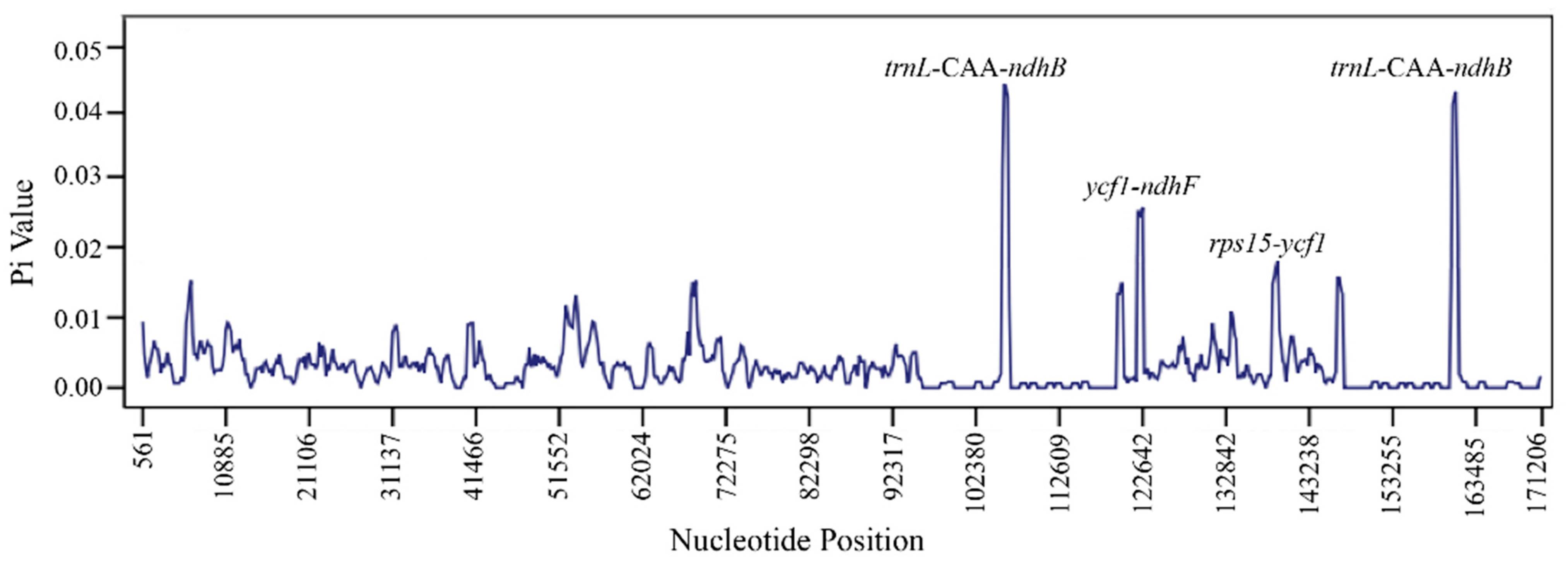
3.3. Repeats, Nucleotide Diversity, Codon Usage and RNA Editing Sites
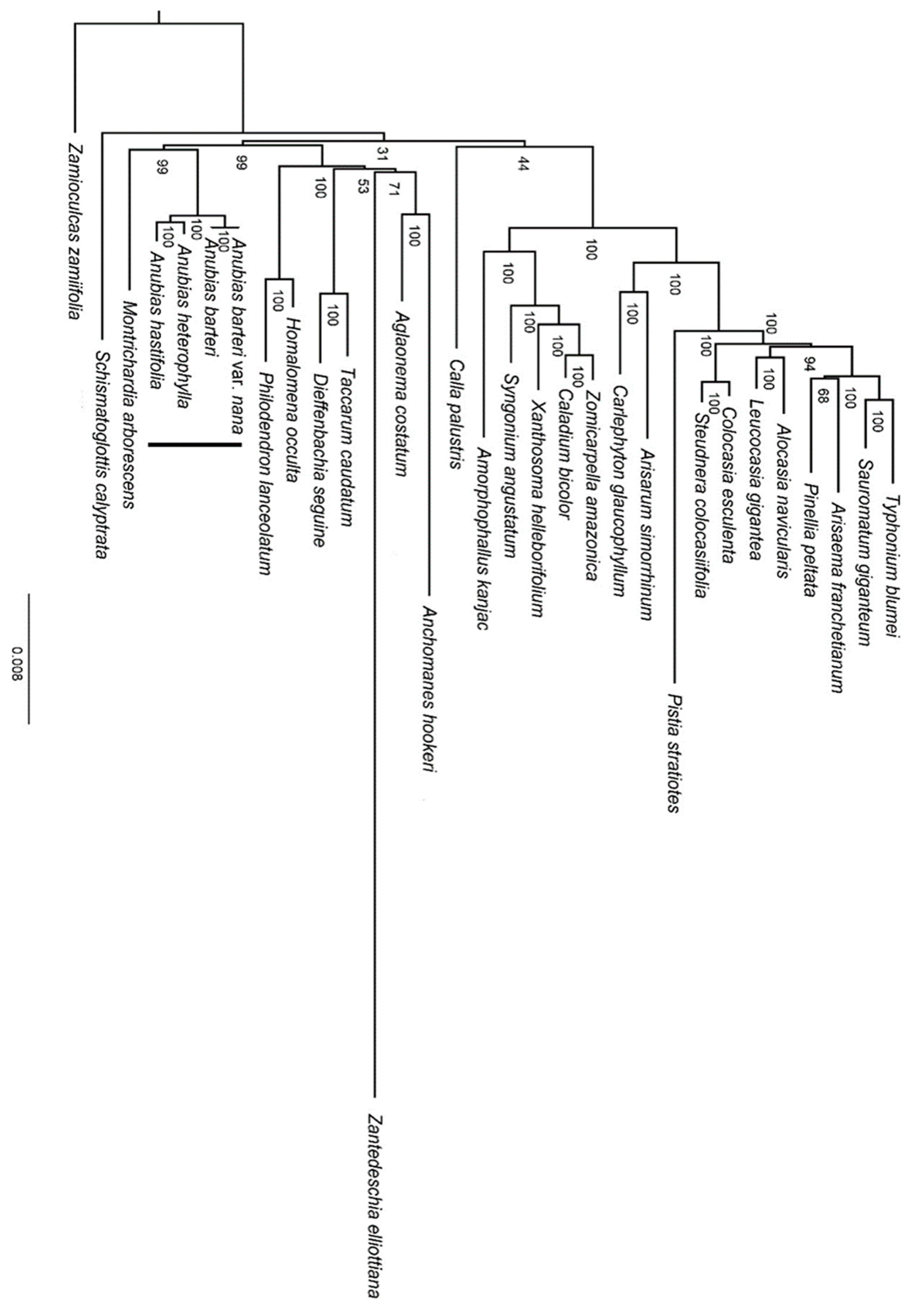
3.4. Phylogenetic Analysis
3.5. Selective Pressure Analysis
4. Discussion
4.1. Chloroplast Genome Features and Comparisons
4.2. Phylogenetic Inference
4.3. Adaptations to the Aquatic Environment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crusio, W.E. A revision of Anubias Schott (Araceae). Meded. Landbouwhogesch. 1979, 79, 1–48. [Google Scholar]
- Mayo, S.; Bogner, J.; Boyce, P. The Genera of Araceae; Royal Botanical Gardens, Kew: London, UK, 1997; pp. 180–182. [Google Scholar]
- Cook, C. The number and kinds of embryo-bearing plants which have become aquatic: A survey. Perspect. Plant Ecol. Evol. Syst. 1999, 2, 79–102. [Google Scholar] [CrossRef]
- Saad, M.; Sahidin, N.; Bhassu, S. DNA Barcoding of Selected Aquarium Plants, Anubias Species Using Chloroplast Marker. In Proceedings of the 18th Scientific Meeting Malaysian Society for Molecular Biology and Biotechnology (MSMBB), Kuala Lumpur, Malaysia, 18–20 August 2009. [Google Scholar] [CrossRef]
- Brunel, S. Pathway analysis: Aquatic plants imported in 10 EPPO Countries. EPPO Bull. 2009, 39, 201–213. [Google Scholar] [CrossRef]
- Oyedeji, A.; Abowei, J. The classification, distribution, control and economic importance of aquatic plants. Int. J. Fish. Aquat. Sci. 2012, 1, 118–128. [Google Scholar]
- Sholichah, L.; Yamin, M.; Ginanjar, R.; Meilisza, N. Anubias (Anubias sp.) propagation trough hydroponic culture technique. J. Phys. Conf. Ser. 2020, 1422, 012024. [Google Scholar] [CrossRef]
- Han, S.Q. Study on Ornamental Water Grass Species and Cultivation Technique. Master’s Thesis, Zhejing University, Hangzhou, China, 2005. [Google Scholar]
- Duminil, J.; Di Michele, M. Plant species delimitation: A comparison of morphological and molecular markers. Plant Biosyst. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- Hollingsworth, P.; Li, D.Z.; van der Bank, M.; Twyford, A. Telling plant species apart with DNA: From barcodes to genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150338. [Google Scholar] [CrossRef]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef]
- Henriquez, C.; Abdullah; Ahmed, I.; Carlsen, M.; Zuluaga, A.; Croat, T.; Mckain, M. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics 2020, 112, 2349–2360. [Google Scholar] [CrossRef]
- Chen, L.Y.; Lu, B.; Morales-Briones, D.F.; Moody, M.L.; Liu, F.; Hu, G.W.; Huang, C.H.; Chen, J.M.; Wang, Q.F. Phylogenomic Analyses of Alismatales Shed Light into Adaptations to Aquatic Environments. Mol. Biol. Evol. 2022, 39, msac079. [Google Scholar] [CrossRef]
- Jung, J.; Lee, S.C.; Choi, H.-K. Anatomical patterns of aerenchyma in aquatic and wetland plants. J. Plant Biol. 2008, 51, 428–439. [Google Scholar] [CrossRef]
- Nowak, J.S.; Ono, J.; Cronk, Q.C. Anatomical study of an aquatic mustard: Subularia aquatica (Brassicaceae). Aquat. Bot. 2010, 93, 55–58. [Google Scholar] [CrossRef]
- Billet, K.; Genitoni, J.; Bozec, M.; Renault, D.; Barloy, D. Aquatic and terrestrial morphotypes of the aquatic invasive plant, Ludwigia grandiflora, show distinct morphological and metabolomic responses. Ecol Evol. 2018, 8, 2568–2579. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; de Pamphilis, C.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Qu, X.J.; Moore, M.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Chan, P.; Lowe, T. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Darling, A.; Mau, B.; Blattner, F.; Perna, N. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.; Pachter, L.; Poliakov, A.; Rubin, E.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-Web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sánchez-Gracia, A. DnaSP6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.; Li, W.H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Peden, J. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 1999. [Google Scholar]
- Wright, F. The Effective Number of Codons Used in A Gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Mower, J. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar] [CrossRef]
- Price, M.; Dehal, P.; Arkin, A. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H.; Nielsen, R. Synonymous and Nonsynonymous Rate Variation in Nuclear Genes of Mammals. J. Mol. Evol. 1998, 46, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.L.; Chen, C.J.; Arab, D.; Du, Z.G.; He, Y.H.; Ho, S. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Henriquez, C.; Arias, T.; Pires, J.; Croat, T.; Schaal, B. Phylogenomics of the plant family Araceae. Mol. Phylogenet. Evol. 2014, 75, 91–102. [Google Scholar] [CrossRef]
- Abdullah; Henriquez, C.; Mehmood, F.; Hayat, A.; Sammad, A.; Waseem, S.; Waheed, M.; Matthews, P.; Croat, T.; Poczai, P.; et al. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics 2021, 113, 183–192. [Google Scholar] [CrossRef]
- Wang, R.J.; Cheng, C.L.; Chang, C.C.; Wu, C.L.; Su, T.M.; Chaw, S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36. [Google Scholar] [CrossRef]
- Hu, S.Q.; Li, G.J.; Yang, J.J.; Hou, H.W. Aquatic Plant Genomics: Advances, Applications, and Prospects. Int. J. Genom. 2017, 2017, 6347874. [Google Scholar] [CrossRef]
- Dong, F.; Lin, Z.C.; Lin, J.; Ming, R.; Zhang, W.P. Chloroplast Genome of Rambutan and Comparative Analyses in Sapindaceae. Plants 2021, 10, 283. [Google Scholar] [CrossRef]
- Mcdonald, M.; Wang, W.C.; Huang, H.D.; Leu, J.Y. Clusters of Nucleotide Substitutions and Insertion/Deletion Mutations Are Associated with Repeat Sequences. PLoS Biol. 2011, 9, e1000622. [Google Scholar] [CrossRef]
- Ahmed, I.; Biggs, P.; Matthews, P.; Collins, L.; Hendy, M.; Lockhart, P. Mutational Dynamics of Aroid Chloroplast Genomes. Genome Biol. Evol. 2012, 4, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, S.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- LaBella, A.; Opulente, D.; Steenwyk, J.; Hittinger, C.; Rokas, A. Variation and selection on codon usage bias across an entire subphylum. PLoS Genet. 2019, 15, e1008304. [Google Scholar] [CrossRef]
- Raman, G.; Park, S.; Lee, E.; Park, S. Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Sci. Rep. 2019, 9, 5028. [Google Scholar] [CrossRef]
- Maier, R.; Zeltz, P.; Kössel, H.; Bonnard, G.; Gualberto, J.; Grienenberger, J. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996, 32, 343–365. [Google Scholar] [CrossRef]
- Wang, M.X.; Liu, H.; Ge, L.Q.; Xing, G.W.; Wang, M.; Song, W.N.; Nie, X.J. Identification and Analysis of RNA Editing Sites in the Chloroplast Transcripts of Aegilops tauschii L. Genes 2016, 8, 13. [Google Scholar] [CrossRef]
- Hoch, B.; Maier, R.; Appel, K.; Igloi, G.; Kössel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 1991, 353, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell. Mol. Life Sci. 2006, 63, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G. Cytology and systematics of Araceae. Nord. J. Bot. 1989, 9, 119–166. [Google Scholar] [CrossRef]
- Săndulescu, E.B.; Stavrescu-Bedivan, M.M.; Vasile Scăețeanu, G.; Șchiopu, T. Morpho-Anatomic features and chemical compounds in some aquatic plant species—Preliminary data. In Proceedings of the Agro for Life. Life for Agriculture. Scientific Papers. Series A. Agronomy, Bucharest, Romania, 5–7 June 2014; Volume LVII, pp. 441–447. [Google Scholar] [CrossRef]
- Schulze, E.-D.; Beck, E.; Buchmann, N.; Clemens, S.; Müller-Hohenstein, K.; Scherer-Lorenzen, M. Plant Ecology, 2nd ed.; Springer: Berlin, Germany, 2019; pp. 143–162. [Google Scholar] [CrossRef]
- Peredo, E.L.; King, U.M.; Les, D.H. The Plastid Genome of Najas Flexilis: Adaptation to Submersed Environments Is Accompanied by the Complete Loss of the NDH Complex in an Aquatic Angiosperm. PLoS ONE 2013, 8, e68591. [Google Scholar] [CrossRef]
- Chen, J.; Zang, Y.; Shang, S.; Liang, S.; Zhu, M.; Wang, Y.; Tang, X. Comparative Chloroplast Genomes of Zosteraceae Species Provide Adaptive Evolution Insights into Seagrass. Front. Plant Sci. 2021, 12, 741152. [Google Scholar] [CrossRef]
- Jiang, B.; Gao, L.; Li, J.; Zhou, Y.; Su, Y.J.; Wang, T. Adaptive evolution of the chloroplast genome in AA-genome Oryza species. Chin. Sci. Bull. 2014, 59, 1975–1983. [Google Scholar] [CrossRef]
- Gu, C.H.; Ma, L.; Wu, Z.Q.; Chen, K.; Wang, Y.X. Comparative analyses of chloroplast genomes from 22 Lythraceae species: Inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 2019, 19, 281. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.Y.; Merchant, S. The Plastid-encoded ccsA Gene Is Required for Heme Attachment to Chloroplast c-type Cytochromes. J. Biol. Chem. 1996, 271, 4632–4639. [Google Scholar] [CrossRef] [PubMed]
- Mavridou, D.; Ferguson, S.; Stevens, J. Cytochrome c assembly. IUBMB Life 2013, 65, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.W.; Zhao, Y.L.; Li, C.Y.; Yang, G.Y.; Peng, J.; Xu, Z.G. New insights into the evolutionary characteristic between the New World and Old World Lupinus species using complete chloroplast genomes. All Life 2021, 14, 414–427. [Google Scholar] [CrossRef]
- Barthet, M.; Hilu, K. Expression of matK: Functional and evolutionary implications. Am. J. Bot. 2007, 94, 1402–1412. [Google Scholar] [CrossRef]
- Zoschke, R.; Nakamura, M.; Liere, K.; Sugiura, M.; Börner, T.; Schmitz-Linneweber, C. An Organellar Maturase Associates with Multiple Group II Introns. PNAS 2010, 107, 3245–3250. [Google Scholar] [CrossRef]
- Hertel, S.; Zoschke, R.; Neumann, L.; Qu, Y.J.; Axmann, I.; Schmitz-Linneweber, C. Multiple Checkpoints for the Expression of the Chloroplast-Encoded Splicing Factor MatK. Plant Physiol. 2013, 163, 1686–1698. [Google Scholar] [CrossRef]
- Wang, R.N.; Milne, R.; Du, X.Y.; Liu, J.; Wu, Z.Y. Characteristics and Mutational Hotspots of Plastomes in Debregeasia (Urticaceae). Front. Genet. 2020, 11, 729. [Google Scholar] [CrossRef]
- Li, J.J.; Liu, H.X.; Mao, S.Z.; Zhao, B.; Huang, S.X. Adaptive evolution of the ndhF gene in the genus Rheum (Polygonaceae). Guihaia 2016, 36, 101–106. [Google Scholar]
- Darshetkar, A.M.; Maurya, S.; Lee, C.; Bazarragchaa, B.; Batdelger, G.; Janchiv, A.; Jeong, E.J.; Choi, S.; Choudhary, R.K.; Kim, S.-Y. Plastome analysis unveils Inverted Repeat (IR) expansion and positive selection in Sea Lavenders (Limonium, Plumbaginaceae, Limonioideae, Limonieae). PhytoKeys 2021, 175, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.-I.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004, 429, 579–582. [Google Scholar] [CrossRef]
- Rumeau, D.; Becuwe-Linka, N.; Beyly, A.; Louwagie, M.; Garin, J.; Peltier, G. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 2005, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Li, D.M.; Li, J.; Wang, D.R.; Xu, Y.C.; Zhu, G.F. Molecular evolution of chloroplast genomes in subfamily Zingiberoideae (Zingiberaceae). BMC Plant Biol. 2021, 21, 558. [Google Scholar] [CrossRef]
- Dong, W.L.; Wang, R.N.; Zhang, N.Y.; Fan, W.B.; Fang, M.F.; Li, Z.H. Molecular evolution of chloroplast genomes of orchid species: Insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 716. [Google Scholar] [CrossRef] [PubMed]
- Krech, K.; Ruf, S.; Masduki, F.; Thiele, W.; Bednarczyk, D.; Albus, C.; Tiller, N.; Hasse, C.; Schöttler, M.; Bock, R. The Plastid Genome-Encoded Ycf4 Protein Functions as a Nonessential Assembly Factor for Photosystem I in Higher Plants. Plant Physiol. 2012, 159, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Dogra, V.; Duan, J.; Lee, K.; Kim, C. Impaired PSII proteostasis triggers an UPR-like response in the var2 mutant of Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 3075–3088. [Google Scholar] [CrossRef]


| Category of Genes | Group of Gene | Name of Gene |
|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4.5 ×2, rrn5 ×2, rrn16 ×2, rrn23 ×2 |
| Transfer RNA genes | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU *, trnL-UAA *, trnL-UAG, trnM, trnM-CAU ×2, trnP-UGG, trnQ-UUG, trnR-UCU, trnS-CGA, trnS-GCU, trnS-GGA, trnS-UGA, trnT-UGU, trnT-GGU, trnV-UAC *, trnY-GUA, trnW-CCA, trnfM-CAU, trnA-UGC *,×2, trnI-GAU *,×2, trnL-CAA ×2, trnN-GUU ×2, trnR-ACG ×2, trnV-GAC ×2 | |
| Ribosomal protein (small subunit) | rps2, rps3, rps4, rps7×2, rps8, rps11, rps12 **,×2, rps14, rps15, rps16 *, rps18, rps19 | |
| Ribosomal protein (large subunit) | rpl2 *,×2, rpl14, rpl16 *, rpl20, rpl22, rpl23 ×2, rpl32, rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Translational initiation factor | infA | |
| Genes for photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3 **, ycf4 |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of cytochrome | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of Rubisco | rbcL | |
| Subunits of NADH dehydrogenase | ndhA *, ndhB *,×2, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Other genes | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA | accD | |
| Synthesis gene | ccsA | |
| ATP-dependent protease | clpP ** | |
| Component of TIC complex | ycf1×2 | |
| Genes of unknown function | Conserved open reading frames | ycf2×2 |
| Gene Name | Positive Sites |
|---|---|
| ccsA | 90 Y 0.935, 91 F 0.843, 92 R 0.636, 198 H 0.937, 211 Y 0.598, 216 L 0.625 |
| matK | 37 L 0.630, 46 E 0.618, 94 F 0.607, 95 D 0.933, 214 R 0.660, 393 P 0.615, 459 P 0.936 |
| ndhF | 49 N 0.579, 53 V 0.566, 291 M 0.568, 463 Q 0.575, 571 D 0.568, 644 G 0.871 |
| ycf4 | 129 G 0.886 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Liu, C.; Hou, K.; Liu, W. Comparative Analyses of Plastomes of Four Anubias (Araceae) Taxa, Tropical Aquatic Plants Endemic to Africa. Genes 2022, 13, 2043. https://doi.org/10.3390/genes13112043
Li L, Liu C, Hou K, Liu W. Comparative Analyses of Plastomes of Four Anubias (Araceae) Taxa, Tropical Aquatic Plants Endemic to Africa. Genes. 2022; 13(11):2043. https://doi.org/10.3390/genes13112043
Chicago/Turabian StyleLi, Li, Changkun Liu, Kunpeng Hou, and Wenzhe Liu. 2022. "Comparative Analyses of Plastomes of Four Anubias (Araceae) Taxa, Tropical Aquatic Plants Endemic to Africa" Genes 13, no. 11: 2043. https://doi.org/10.3390/genes13112043
APA StyleLi, L., Liu, C., Hou, K., & Liu, W. (2022). Comparative Analyses of Plastomes of Four Anubias (Araceae) Taxa, Tropical Aquatic Plants Endemic to Africa. Genes, 13(11), 2043. https://doi.org/10.3390/genes13112043





