Retinal Gene Expression of Selective Genes and Histological Stages of Embryonic and Post-Hatch Chickens (Gallus gallus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animal Source for the Study
2.3. Histology
2.4. RNA Extraction and cDNA Synthesis
2.5. RT-PCR
2.6. Sanger Sequencing
2.7. RT-qPCR
3. Results
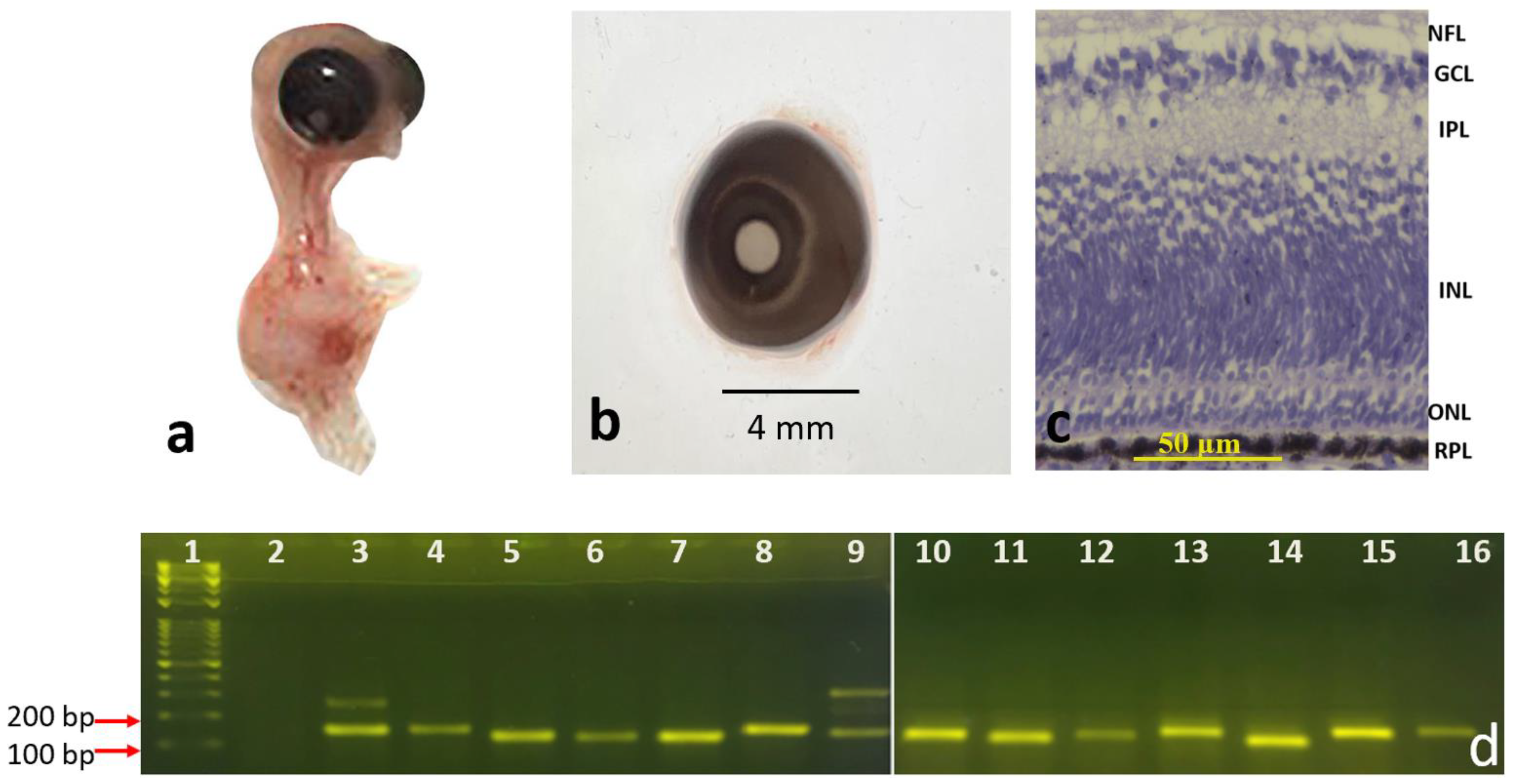
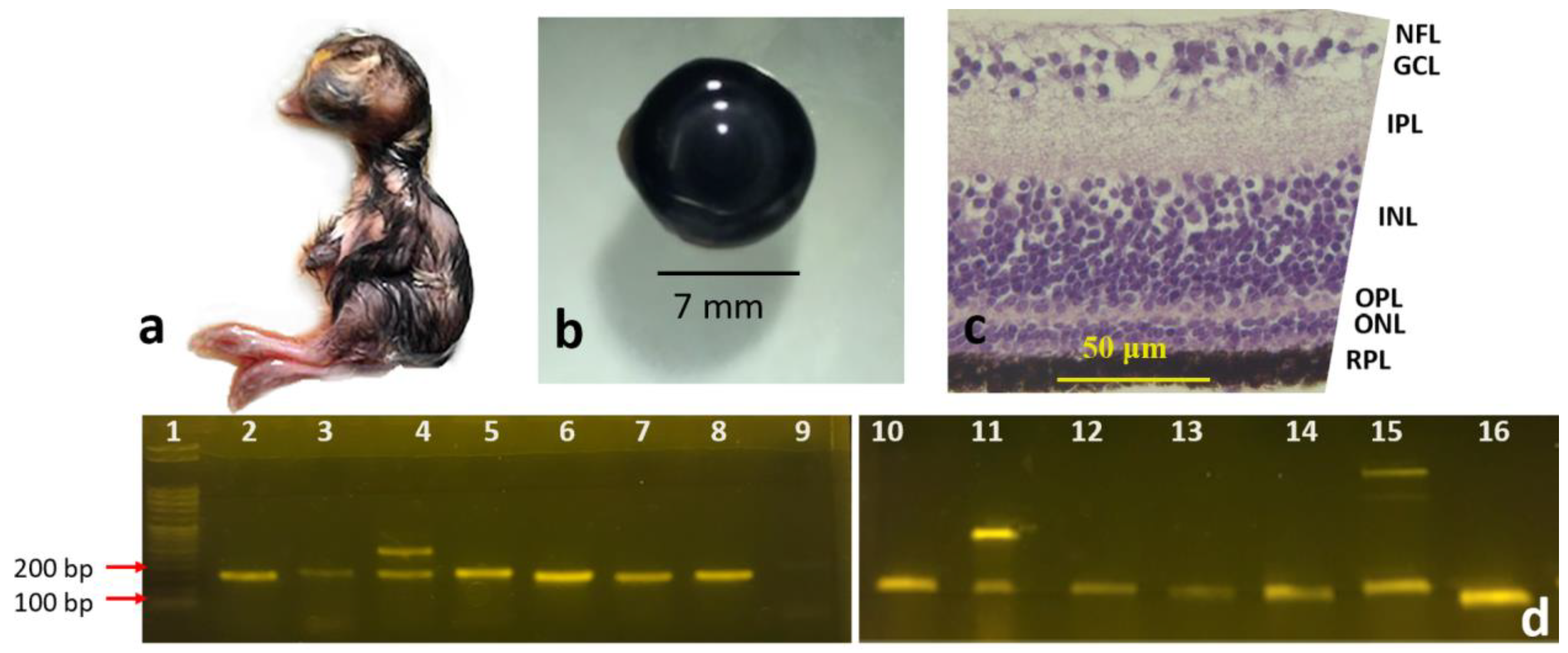
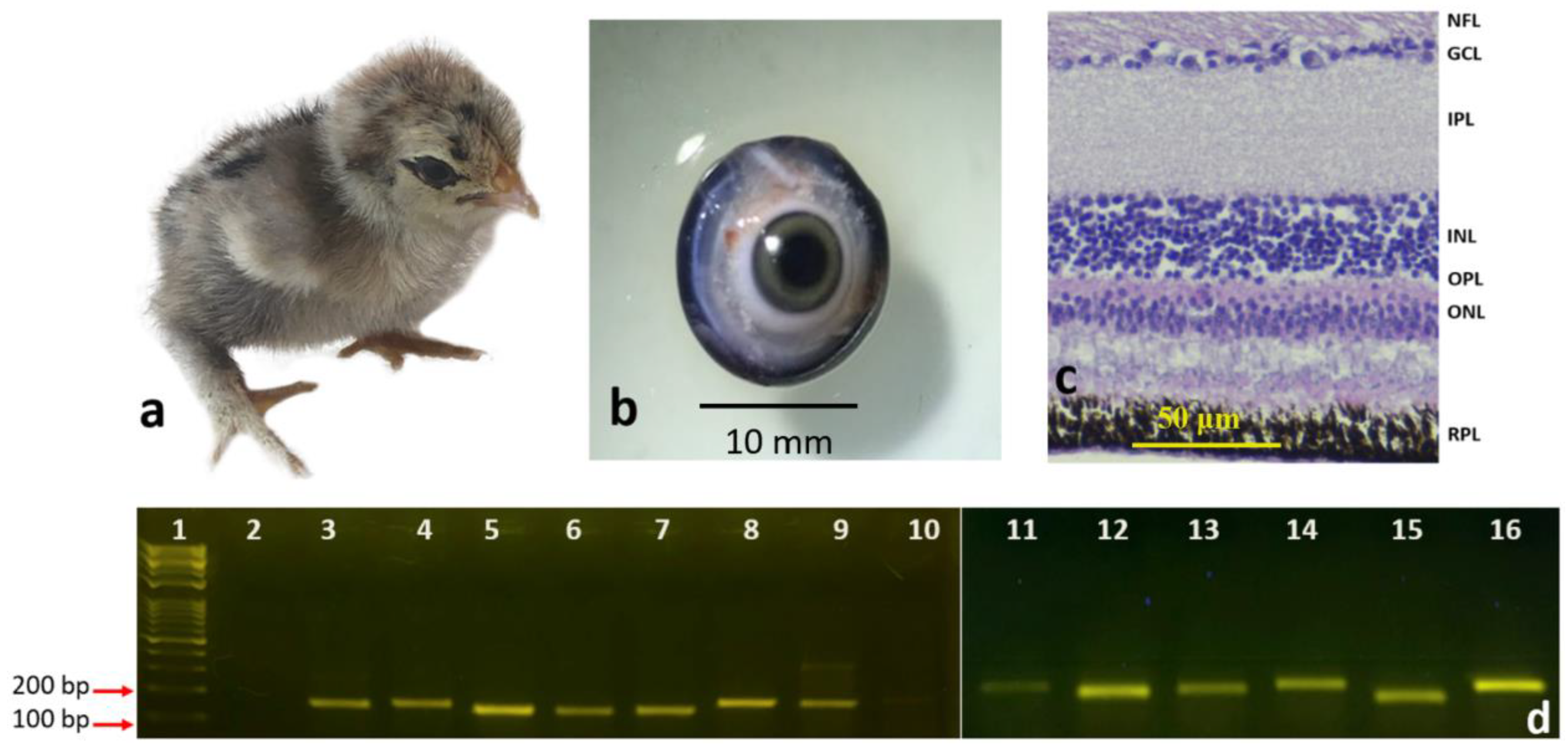
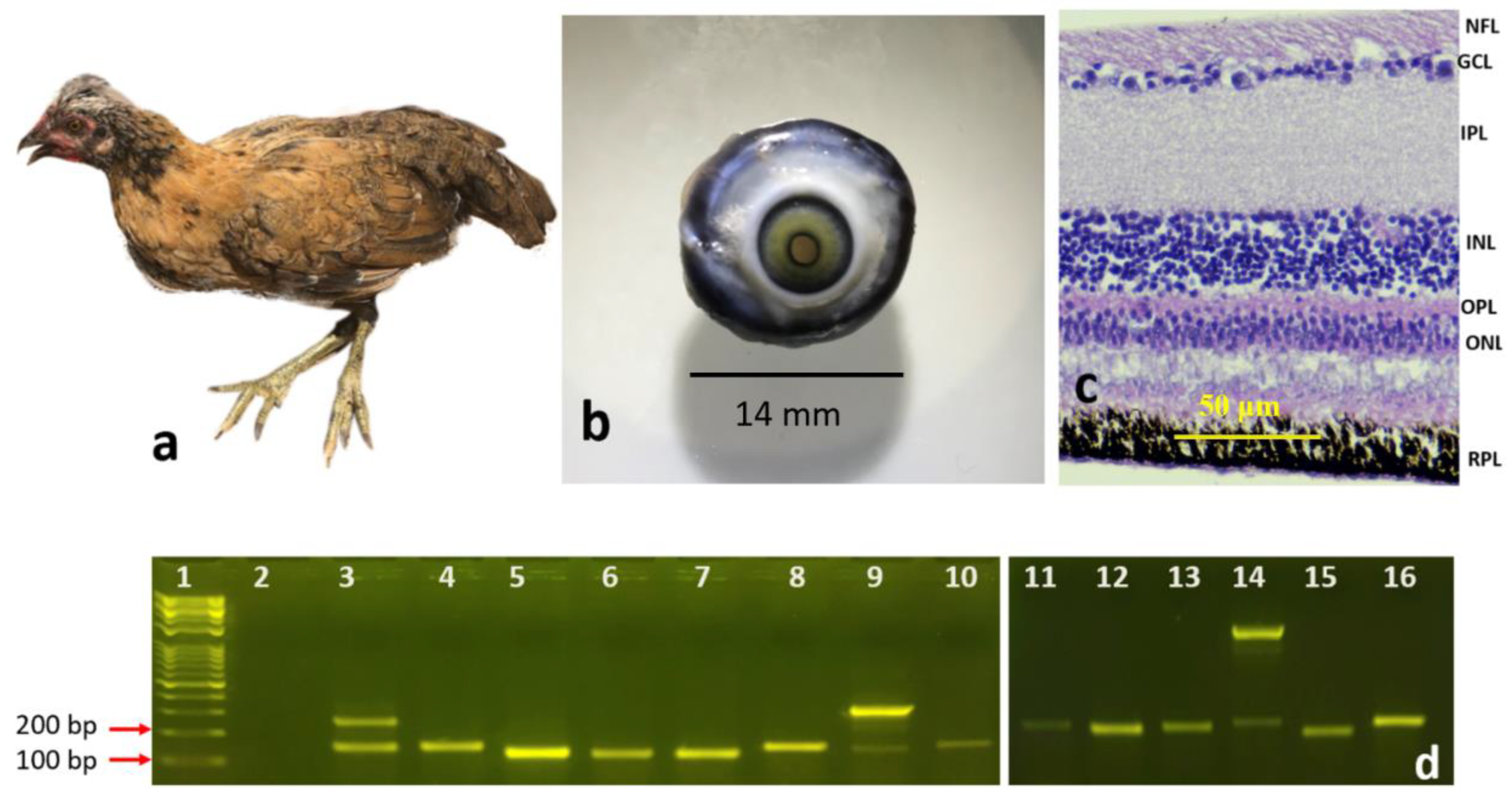
3.1. Morphological Differentiation Stages of the Developing Retinal Layers
3.1.1. Eight-Day and Fifteen-Day Chick Embryos
3.1.2. Newly Hatched Chick and the Adult Chicken
3.2. Gene Expression Analysis Using RT-PCR
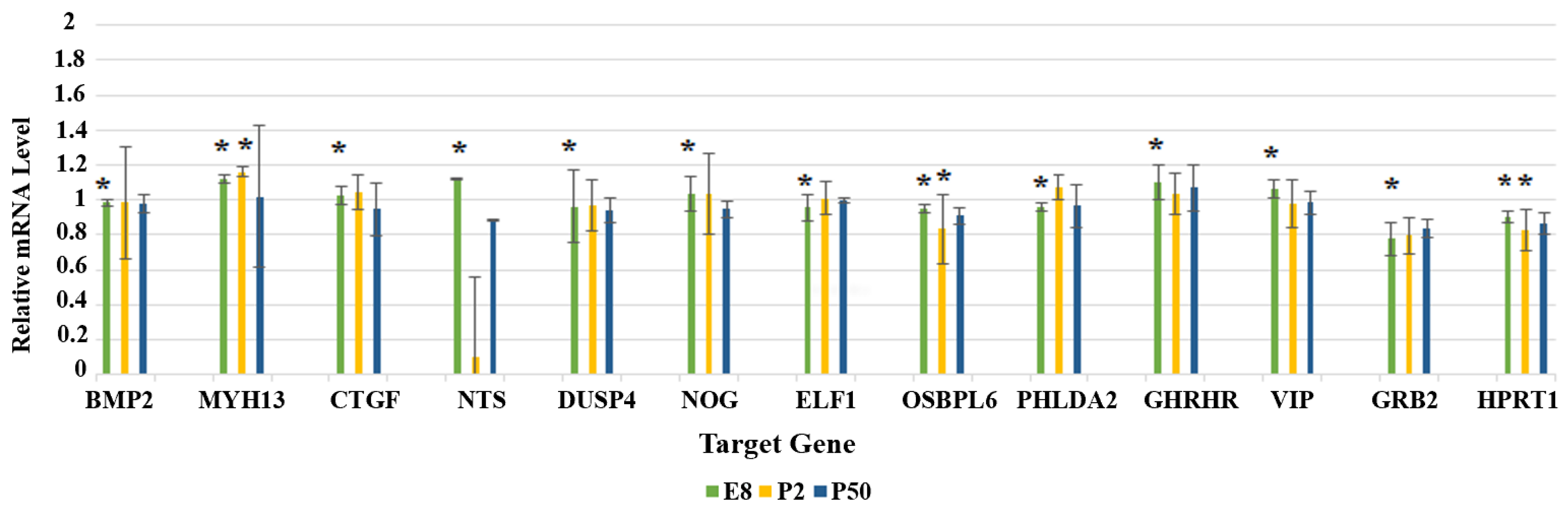
3.3. Gene Expression Analysis Using RT-qPCR
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- da Silva, S.; Cepko, C.L. Fgf8 expression, and degradation of retinoic acid are required for patterning a high-acuity area in the retina. Dev. Cell 2017, 42, 68–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lev, S. Molecular aspects of retinal degenerative diseases. Cell Mol. Neurobiol. 2001, 21, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Baden, T.; Osorio, D. The retinal basis of vision in chicken. Semin. Cell Dev. Biol. 2020, 106, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Langouet-Astrie, C.J.; Meinsen, A.L.; Grunwald, E.R.; Turner, S.D.; Enke, R.A. RNA sequencing analysis of the developing chicken retina. Sci. Data 2016, 3, 160117. [Google Scholar] [CrossRef] [PubMed]
- Wisely, C.E.; Sayed, J.A.; Tamez, H.; Zelinka, C.; Abdel-Rahman, M.H.; Fischer, A.J.; Cebulla, C.M. The chick eye in vision research: An excellent model for the study of ocular disease. Prog. Retin. Eye Res. 2017, 61, 72–97. [Google Scholar] [CrossRef]
- Adler, R.; Raymond, P.A. Have we achieved a unified model of photoreceptor cell fate specification in vertebrates? Brain Res. 2008, 1192, 134–150. [Google Scholar] [CrossRef] [Green Version]
- Haider, N.B.; Ikeda, A.; Naggert, J.K.; Nishina, P.M. Genetic modifiers of vision and hearing. Hum. Mol. Genet. 2002, 11, 1195–1206. [Google Scholar] [CrossRef] [Green Version]
- McGlinn, A.M.; Baldwin, D.A.; Tobias, J.W.; Budak, M.T.; Khurana, T.S.; Stone, R.A. Form-deprivation myopia in chick induces limited changes in retinal gene expression. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3430–3436. [Google Scholar] [CrossRef]
- Schippert, R.; Schaeffel, F.; Feldkaemper, M.P. Microarray analysis of retinal gene expression in chicks during imposed myopic defocus. Mol. Vis. 2008, 14, 1589–1599. [Google Scholar]
- Stone, R.A.; McGlinn, A.M.; Baldwin, D.A.; Tobias, J.W.; Iuvone, P.M.; Khurana, T.S. Image defocus and altered retinal gene expression in chick: Clues to the pathogenesis of ametropia. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5765–5777. [Google Scholar] [CrossRef]
- Stone, R.A.; Khurana, T.S. Gene profiling in experimental models of eye growth: Clues to myopia pathogenesis. Vis. Res. 2010, 50, 2322–2333. [Google Scholar] [CrossRef]
- Rocha-Martins, M.; Njaine, B.; Silveira, M.S. Avoiding pitfalls of internal controls: Validation of reference genes for analysis by qRT-PCR and Western blot throughout rat retinal development. PLoS ONE 2012, 7, e43028. [Google Scholar] [CrossRef]
- Moon, S.; Lee, S.; Caesar, J.A.; Pruchenko, S.; Leask, A.; Knowles, J.A.; Sinon, J.; Chaqour, B. A CTGF-YAP regulatory pathway is essential for angiogenesis and barriergenesis in the retina. iScience 2020, 23, 101184. [Google Scholar] [CrossRef]
- National Research Council. Guide for the Care and Use of Laboratory Animals, 8th ed.; The National Academies Press: Washington, DC, USA, 1996.
- Zareen, N.; Khan, M.Y.; Minhas, L.A. Histological stages of retinal morphogenesis in chicken—A descriptive laboratory research. Ital. J. Zool. 2011, 78, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Hang, A.; Phan, E.; Wildsoet, C.F. Differential gene expression of BMP2 and BMP receptors in chick retina & choroid induced by imposed optical defocus. Vis. Neurosci. 2016, 33, E015. [Google Scholar] [CrossRef] [Green Version]
- Riddell, N.; Giummarra, L.; Hall, N.E.; Crewther, S.G. Bidirectional expression of metabolic, structural, and immune pathways in early myopia and hyperopia. Front. Neurosci. 2016, 10, 390. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.S.; Lee, W.K.; Moon, Y.S.; Kim, N.R. Early changes in retinal structure and BMP2 expression in the retina and crystalline lens of streptozotocin-induced diabetic pigs. Lab. Anim. Res. 2017, 33, 216–222. [Google Scholar] [CrossRef] [Green Version]
- Elmasry, K.; Habib, S.; Moustafa, M.; Al-Shabrawey, M. Bone morphogenetic proteins and diabetic retinopathy. Biomolecules 2021, 11, 593. [Google Scholar] [CrossRef]
- Lazarev, V.F.; Guzhova, I.V.; Margulis, B.A. Glyceraldehyde-3-phosphate dehydrogenase is a multifaceted therapeutic target. Pharmaceutics 2020, 12, 416. [Google Scholar] [CrossRef]
- Ma, C.; Zhong, P.; Liu, D.; Barger, Z.K.; Zhou, L.; Chang, W.C.; Kim, B.; Dan, Y. Sleep regulation by neurotensinergic neurons in a thalamo-amygdala circuit. Neuron 2019, 103, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Devader, C.; Béraud-Dufour, S.; Coppola, T.; Mazella, J. The anti-apoptotic role of neurotensin. Cells 2013, 2, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Pi, L.; Xia, H.; Liu, J.; Shenoy, A.K.; Hauswirth, W.W.; Scott, E.W. Role of connective tissue growth factor in the retinal vasculature during development and ischemia. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8701–8710. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.J.; Pi, L.; Sriram, S.; Mao, C.; Petersen, B.E.; Scott, E.W.; Leask, A.; Schultz, G.S. Conditional knockout of CTGF affects corneal wound healing. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2062–2070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, J.A.; Kilbane Myers, J.; Khan, M.; Angelos, M.G.; Chen, C.A. Dual-specificity phosphatase 4 overexpression in cells prevents hypoxia/reoxygenation-induced apoptosis via the upregulation of eNOS. Front. Cardiovasc. Med. 2017, 4, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cakmak, A.I.; Basmak, H.; Gursoy, H.; Ozkurt, M.; Yildirim, N.; Erkasap, N.; Bilgec, M.D.; Tuncel, N.; Colak, E. Vasoactive intestinal peptide, a promising agent for myopia? Int. J. Ophthalmol. 2017, 10, 211–216. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Ho, C.; Hammond, D.; Wildsoet, C.F. Differential expression of BMP7, TGF-β2, and Noggin in chick RPE after imposed optical defocus. Investig. Ophthalmol. Vis. Sci. 2012, 53, 3458. [Google Scholar]
- Messina, A.; Incitti, T.; Bozza, A.; Bozzi, Y.; Casarosa, S. Noggin expression in the adult retina suggests a conserved role during vertebrate evolution. J. Histochem. Cytochem. 2014, 62, 532–540. [Google Scholar] [CrossRef] [Green Version]
- Brandli, A.; Gerhart, J.; Sutera, C.K.; Purushothuman, S.; George-Weinstein, M.; Stone, J.; Bravo-Nuevo, A. Role of Myo/Nog cells in neuroprotection: Evidence from the light damaged retina. PLoS ONE 2017, 12, e0169744. [Google Scholar] [CrossRef] [Green Version]
- Moreira, E.F.; Jaworski, C.; Li, A.; Rodriguez, I.R. Molecular and biochemical characterization of a novel oxysterol-binding protein (OSBP2) highly expressed in retina. J. Biol. Chem. 2001, 276, 18570–18578. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.A.; Karabina, A.; Broadwell, L.J.; Leinwand, L.A. The ancient sarcomeric myosins found in specialized muscles. Skelet. Muscle 2019, 9, 7. [Google Scholar] [CrossRef] [Green Version]
- Rajala, R.V.; Anderson, R.E. Light regulation of the insulin receptor in the retina. Mol. Neurobiol. 2003, 28, 123–138. [Google Scholar] [CrossRef]
- Burdon, K.P.; Fogarty, R.D.; Shen, W.; Abhary, S.; Kaidonis, G.; Appukuttan, B.; Hewitt, A.W.; Sharma, S.; Daniell, M.; Essex, R.W.; et al. Genome-wide association study for sight-threatening diabetic retinopathy reveals association with genetic variation near the GRB2 gene. Diabetologia 2015, 58, 2288–2297. [Google Scholar] [CrossRef]
- van Wijngaarden, P.; Brereton, H.M.; Coster, D.J.; Williams, K.A. Stability of housekeeping gene expression in the rat retina during exposure to cyclic hyperoxia. Mol. Vis. 2007, 13, 1508–1515. [Google Scholar]
- Liu, X.; Xie, J.; Liu, Z.; Gong, Q.; Tian, R.; Su, G. Identification and validation of reference genes for quantitative RT-PCR analysis of retinal pigment epithelium cells under hypoxia and/or hyperglycemia. Gene 2016, 580, 41–46. [Google Scholar] [CrossRef]
- Ceballos-Picot, I.; Mockel, L.; Potier, M.C.; Dauphinot, L.; Shirley, T.L.; Torero-Ibad, R.; Fuchs, J.; Jinnah, H.A. Hypoxanthine-guanine phosphoribosyl transferase regulates early developmental programming of dopamine neurons: Implications for Lesch-Nyhan disease pathogenesis. Hum. Mol. Genet. 2009, 18, 2317–2327. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, M.; Cheng, H.J.; Friedman, G.C.; Yoon, C.H.; O’Leary, D.D.M. Topographically specific effects of ELF-1 on retinal axon guidance in vitro and retinal axon mapping in vivo. Cell 1996, 86, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.K.; Law, K.S.; Chan, S.O.; Yam, J.C.; Chen, L.J.; Zhang, H.; Cheung, H.S.; Block, N.L.; Schally, A.V.; Pang, C.P. Antagonists of growth hormone-releasing hormone receptor induce apoptosis specifically in retinoblastoma cells. Proc. Natl. Acad. Sci. USA 2016, 113, 14396–14401. [Google Scholar] [CrossRef] [Green Version]
- Thounaojam, M.C.; Powell, F.L.; Patel, S.; Gutsaeva, D.R.; Tawfik, A.; Smith, S.B.; Nussbaum, J.; Block, N.L.; Martin, P.M.; Schally, A.V.; et al. Protective effects of agonists of growth hormone-releasing hormone (GHRH) in early experimental diabetic retinopathy. Proc. Natl. Acad. Sci. USA 2017, 114, 13248–13253. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Valle, M.L.; Beveridge, C.; Liu, Y.; Sharma, S. Unraveling the role of genetics in the pathogenesis of diabetic retinopathy. Eye 2019, 33, 534–541. [Google Scholar] [CrossRef] [Green Version]
- Perera, N.; Ritchie, R.H.; Tate, M. The role of bone morphogenetic proteins in diabetic complications. ACS. Pharmacol. Transl. Sci. 2020, 3, 11–20. [Google Scholar] [CrossRef]
- Madsen-Bouterse, S.; Mohammad, G.; Kowluru, R.A. Glyceraldehyde-3-phosphate dehydrogenase in retinal microvasculature: Implications for the development and progression of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2010, 51, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
| Gallus Gene Full Name | Gene Abbreviation | Validation with RT-qPCR [Reference] |
|---|---|---|
| BMP2 | Bone Morphogenetic Protein 2 | Yes [8,10] |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase | Yes [12,13] |
| NTS | Neurotensin | Not confirmed before this study |
| PHLDA2 | Pleckstrin Homology Like Domain family A member 2 | Not confirmed before this study |
| CTGF | Connective Tissue Growth Factor | Yes [8,13] |
| DUSP4 | Dual Specificity Phosphatase 4 | Yes [10] |
| VIP | Vasoactive Intestinal Peptide | Yes [8,10] |
| NOG | Noggin | Yes [10] |
| OSBPL6 | OxySterol Binding Protein-Like 6 | Yes [9,10] |
| MYH13 | MYosin, Heavy chain 13, skeletal muscle | Not confirmed before this study |
| GRB2 | Growth factor Receptor Bound protein 2 | Yes [9] |
| HPRT1 | Hypoxanthine Phospho-Ribosyl Transferase 1 | Yes [12] |
| ELF1 | E74 Like ETS transcription Factor 1 | Yes [9] |
| GHRHR | Growth Hormone Releasing Hormone Receptor | Yes [9] |
| Gene Abbreviation | NCBI Code (mRNA) | Primers Sequence (5′–3′) F: Forward R: Reverse | PCR Fragment Length (bp) |
|---|---|---|---|
| BMP2 | NM_204358.1 | F: GCCAGAAACAAGTGGGAAAA R: TACGGTGATGGTAGCTGCTG | 149 bp |
| GAPDH | NM_204305.1 | F: CACACAGAAGACGGTGGATG R: CAGCTCAGGGATGACTTTCC | 124 bp |
| NTS | NM_001277360.1 | F: AAGACAGTTCCCTGCTGCTC R: GATCAAATGCGTCTTGCTGA | 128 bp |
| PHLDA2 | NM_001199595.1 | F: GCCATCCAGGACTTCAAGAG R: CCTGTCCCTTTTCAGTCCAA | 144 bp |
| CTGF | NM_204274.1 | F: TTCCAGAGCAGCTGCAAGTA R: ACCCACTCCTCACAGCACTT | 149 bp |
| DUSP4 | NM_204838.1 | F: TGCCTCAGAGTATCCCGAAT R: AAGGGAAGGATTTCCACAGG | 147 bp |
| VIP | NM_205366.2 | F: AAATCCAGCCAAACTTCGAG R: GGTGTCCTTCAGAGGTCCAA | 119 bp |
| NOG | NM_204123.1 | F: CTCGGGGTAGACGATCTGG R: CAGCTTCTTGCTCAGCCTGT | 138 bp |
| OSBPL6 | XM_421982.5 | F: CGAAGATGAGTTCGGAGGAG R: AGCCTTCCGTTCCAAAAGAT | 134 bp |
| MYH13 | XM_015295194. | F: CTGCGGATATCGAAACCTGT R: TAGGGGTTGGTCGAGATGAG | 148 bp |
| GRB2 | NM_204411.1 | F: ATGTGCAGCAGTTCAAGGTG R: GCTGGTTCCTGGAGACAGAT | 123 bp |
| HPRT1 | NM_204848.1 | F: AGCCCCATCGTCATATGCT R: AGCCCCATCGTCATATGCT | 155 bp |
| ELF1 | NM_001006269.1 | F: GGAAATGCCAAAAGATCTCG R: TCCTCCCTTTGTTCCTGTGT | 155 bp |
| GHRHR | NM_001037834.1 | F: CTACGCTGCCCCAGAAATTA R: AAAGCTGCAATGGTCAATGTC | 123 bp |
| Gallus Gene | Function in the Retina or Eye | Disease-Associated-Therapeutic |
|---|---|---|
| BMP2 | Retinal development Retinal growth regulation Embryogenesis [16] | Myopia and hyperopia [16,17] Diabetic retinopathy [18,19] Association with ametropia [10] |
| GAPDH | Housekeeping proteins (glycolysis) [20] | Diabetic retinopathy [20] |
| TS | Retinal neural development [21] Anti-apoptotic role [22] | Association with myopia [8] |
| PHLDA2 | Not listed in the literature | Association with myopia [8] |
| CTGF | Vessel growth during early retinal development [23] | Ischemic retinopathies [23] Angiogenesis [23] Wound healing [24] Myopia [8] |
| DUSP4 | Differentiation Proliferation Survival Apoptosis [25] | Hyperopia [10] |
| VIP | Neuromodulator [26] Stimulation of cell division and differentiation of RPE cells [8] | Myopia [26] |
| NOG | Early eye growth and development of retinal structures [27,28] | Therapeutic in neurodegenerative retinal diseases [29] Association with ametropia [10] |
| OSBPL6 | Neural retina development [30] | Association with ametropia [10] |
| MYH13 | Eye muscle movement and coordination [31] | Association with ametropia [10] |
| GRB2 | Insulin pathway [9] Appropriate functioning of rods (photoreceptor cells) by -insulin pathway [32] Excessive eye growth [9] | Diabetic retinopathy [33] Myopia [9] |
| HPRT1 | Housekeeping gene in the retina [34,35] | Neural development [36] |
| ELF1 | Retinotectal projection [9] ElF1 and its receptors act as retinal exon guidance [37] | Myopia [9] |
| GHRHR | Protective roles in the eye [38] GHRH diminishes retinal neurovascular injury in the early stages of diabetic retinopathy [39] | Retinoblastoma [38] Antioxidant and anti-inflammatory [39] Myopia [9] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastaki, N.K.; Lobo, V.R.; Gomes, T.; Albarjes, T.A. Retinal Gene Expression of Selective Genes and Histological Stages of Embryonic and Post-Hatch Chickens (Gallus gallus). Genes 2022, 13, 2048. https://doi.org/10.3390/genes13112048
Bastaki NK, Lobo VR, Gomes T, Albarjes TA. Retinal Gene Expression of Selective Genes and Histological Stages of Embryonic and Post-Hatch Chickens (Gallus gallus). Genes. 2022; 13(11):2048. https://doi.org/10.3390/genes13112048
Chicago/Turabian StyleBastaki, Nasmah K., Vanessa R. Lobo, Thecla Gomes, and Taybha A. Albarjes. 2022. "Retinal Gene Expression of Selective Genes and Histological Stages of Embryonic and Post-Hatch Chickens (Gallus gallus)" Genes 13, no. 11: 2048. https://doi.org/10.3390/genes13112048
APA StyleBastaki, N. K., Lobo, V. R., Gomes, T., & Albarjes, T. A. (2022). Retinal Gene Expression of Selective Genes and Histological Stages of Embryonic and Post-Hatch Chickens (Gallus gallus). Genes, 13(11), 2048. https://doi.org/10.3390/genes13112048





