From Processivity to Genome Maintenance: The Many Roles of Sliding Clamps
Abstract
:1. Introduction
1.1. Genetic Organization
1.2. Structural Organization
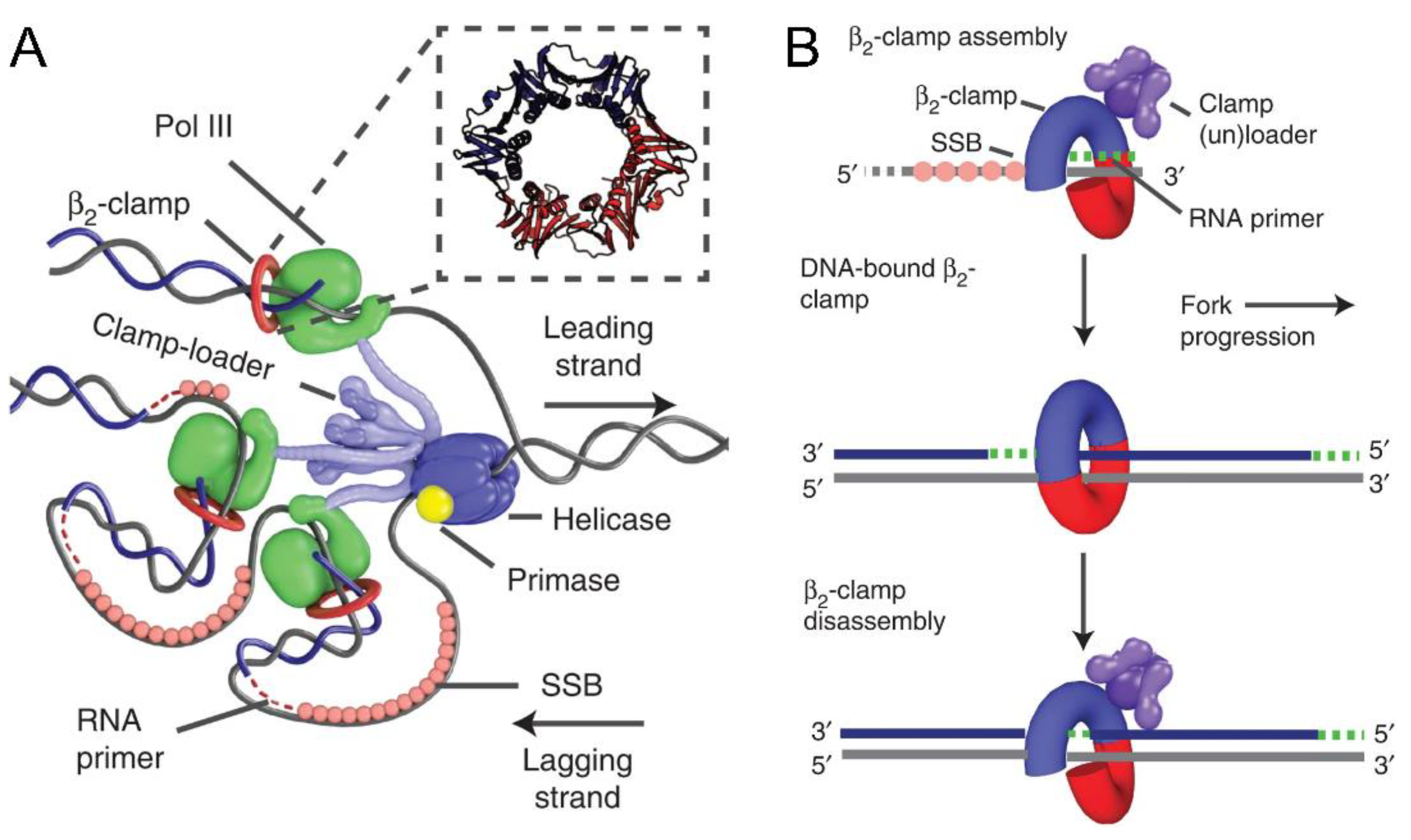
1.3. Sliding Clamps as Processivity Factors
1.4. Beyond the Obvious: Functions Other Than Processivity
1.5. Interactions with Other Members of the Replication Machinery
1.6. Transcription Activation and the Clamp
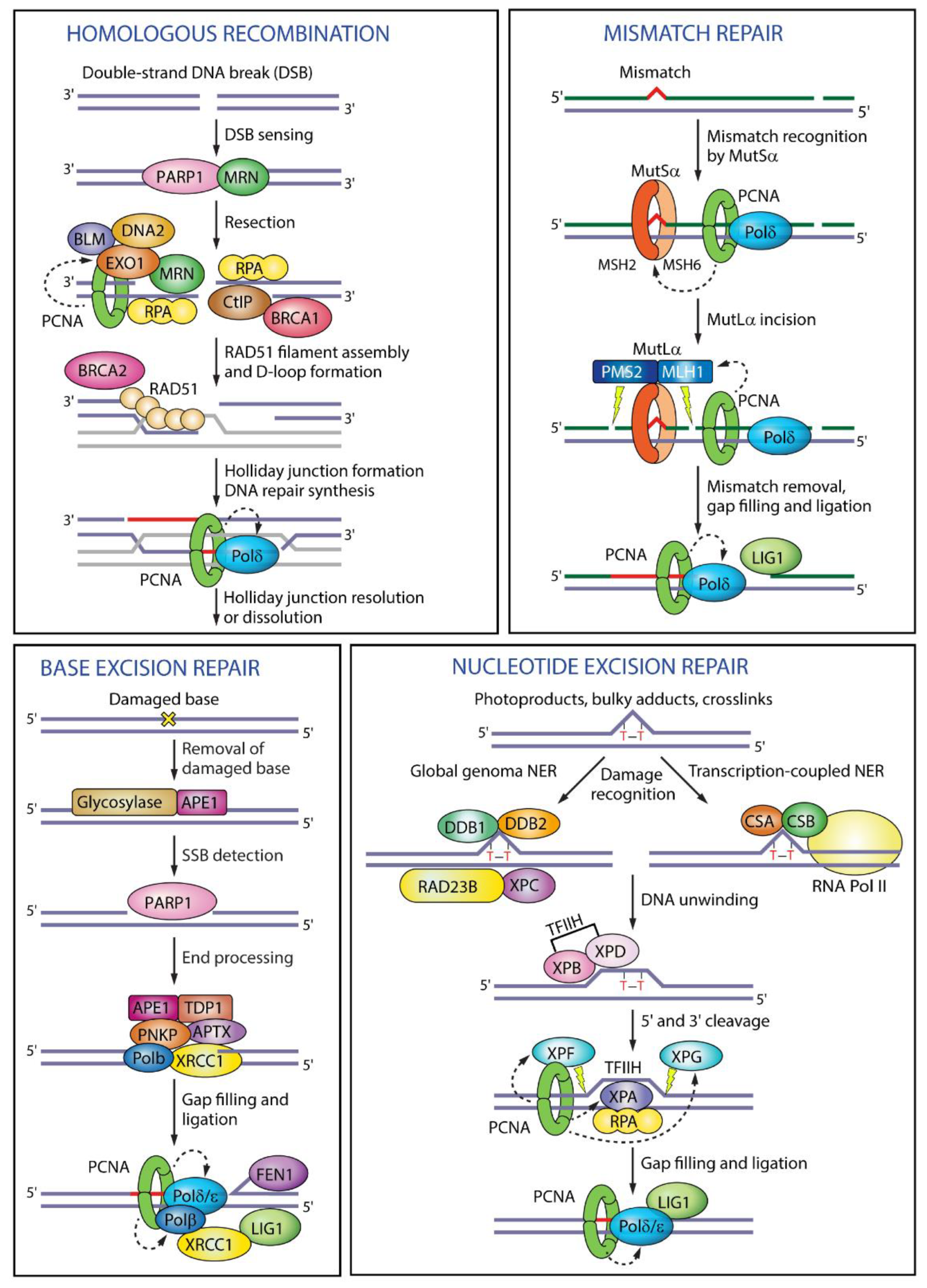
1.7. Role in Maintenance of Genome Integrity
1.8. Translesion Synthesis and the Clamp
1.9. Transposon Interaction with β Clamp
1.10. Other Moonlighting Roles of PCNA
1.11. Unusual Processivity Factors
1.11.1. Viral Clamps
1.11.2. 9-1-1 Clamp
1.11.3. Thioredoxin as the T7 Clamp
1.12. Utilizing the Potential of Clamps: Development as Therapeutic Targets
2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ishino, S.; Ishino, Y. DNA polymerases as useful reagents for biotechnology—The history of developmental research in the field. Front. Microbiol. 2014, 5, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maga, G. DNA Polymerases. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Leman, A.R.; Noguchi, E. The Replication Fork: Understanding the Eukaryotic Replication Machinery and the Challenges to Genome Duplication. Genes 2013, 4, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelman, Z.; O’Donnell, M. DNA replication: Enzymology and mechanisms. Curr. Opin. Genet. Dev. 1994, 4, 185–195. [Google Scholar] [CrossRef]
- Masai, H.; Matsumoto, S.; You, Z.; Yoshizawa-Sugata, N.; Oda, M. Eukaryotic Chromosome DNA Replication: Where, When, and How? Annu. Rev. Biochem. 2010, 79, 89–130. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Ai, Y. Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2010, 1804, 1081–1093. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Hearst, J.; Alberts, B. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J. Biol. Chem. 1981, 256, 4087–4094. [Google Scholar] [CrossRef]
- Kuriyan, J.; O’Donnell, M. Sliding Clamps of DNA Polymerases. J. Mol. Biol. 1993, 234, 915–925. [Google Scholar] [CrossRef]
- Kong, X.-P.; Onrust, R.; O’Donnell, M.; Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 1992, 69, 425–437. [Google Scholar] [CrossRef]
- O’Donnell, M.; Kuriyan, J.; Kong, X.P.; Stukenberg, P.T.; Onrust, R. The sliding clamp of DNA polymerase III holoenzyme encircles DNA. Mol. Biol. Cell 1992, 3, 953–957. [Google Scholar] [CrossRef] [Green Version]
- Georgescu, R.E.; Kim, S.-S.; Yurieva, O.; Kuriyan, J.; Kong, X.-P.; O’Donnell, M. Structure of a Sliding Clamp on DNA. Cell 2008, 132, 43–54. [Google Scholar] [CrossRef]
- Gui, W.-J.; Lin, S.-Q.; Chen, Y.-Y.; Zhang, X.-E.; Bi, L.-J.; Jiang, T. Crystal structure of DNA polymerase III β sliding clamp from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2011, 405, 272–277. [Google Scholar] [CrossRef] [PubMed]
- McGrath, A.E.; Martyn, A.P.; Whittell, L.R.; Dawes, F.E.; Beck, J.L.; Dixon, N.E.; Kelso, M.J.; Oakley, A.J. Crystal structures and biochemical characterization of DNA sliding clamps from three Gram-negative bacterial pathogens. J. Struct. Biol. 2018, 204, 396–405. [Google Scholar] [CrossRef] [Green Version]
- Argiriadi, M.A.; Goedken, E.R.; Bruck, I.; O’Donnell, M.; Kuriyan, J. Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct. Biol. 2006, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, T.S.; Kong, X.-P.; Gary, S.; Burgers, P.M.; Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994, 79, 1233–1243. [Google Scholar] [CrossRef]
- Turner, J.; Hingorani, M.M.; Kelman, Z.; O’Donnell, M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 1999, 18, 771–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruck, I.; O’Donnell, M. The ring-type polymerase sliding clamp family. Genome Biol. 2001, 2, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Perez-Roger, I.; García-Sogo, M.; Navarro-Aviñó, J.; López-Acedo, C.; Macián, F.; Armengod, M. Positive and negative regulatory elements in the dnaA-dnaN-recF operon of Escherichia coli. Biochimie 1991, 73, 329–334. [Google Scholar] [CrossRef]
- Fang, J.; Nevin, P.; Kairys, V.; Venclovas, Č.; Engen, J.R.; Beuning, P.J. Conformational Analysis of Processivity Clamps in Solution Demonstrates that Tertiary Structure Does Not Correlate with Protein Dynamics. Structure 2014, 22, 572–581. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Maganña, A.; Blanco, F.J. Human PCNA Structure, Function and Interactions. Biomolecules 2020, 10, 570. [Google Scholar] [CrossRef] [Green Version]
- Kelch, B.A. Review: The lord of the rings: Structure and mechanism of the sliding clamp loader. Biopolymers 2016, 105, 532–546. [Google Scholar] [CrossRef]
- Ulrich, H.D. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair 2009, 8, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Bellí, G.; Colomina, N.; Castells-Roca, L.; Lorite, N.P. Post-Translational Modifications of PCNA: Guiding for the Best DNA Damage Tolerance Choice. J. Fungi 2022, 8, 621. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-C.; Nakajima, Y.; Yu, Y.-L.; Xia, W.; Chen, C.-T.; Yang, C.-C.; McIntush, E.W.; Li, L.-Y.; Hawke, D.; Kobayashi, R.; et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006, 8, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Maga, G.; Hubscher, U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003, 116, 3051–3060. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.I.; Jain, V. Molecular Dissection of the Homotrimeric Sliding Clamp of T4 Phage: Two Domains of a Subunit Display Asymmetric Characteristics. Biochemistry 2016, 55, 588–596. [Google Scholar] [CrossRef]
- Singh, M.I.; Ganesh, B.; Jain, V. On the domains of T4 phage sliding clamp gp45: An intermolecular crosstalk governs structural stability and biological activity. Biochim. et Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 3300–3310. [Google Scholar] [CrossRef] [PubMed]
- Koleva, B.N.; Gokcan, H.; Rizzo, A.A.; Lim, S.; Fouque, K.J.D.; Choy, A.; Liriano, M.L.; Fernandez-Lima, F.; Korzhnev, D.M.; Cisneros, G.A.; et al. Dynamics of the E. coli β-Clamp Dimer Interface and Its Influence on DNA Loading. Biophys. J. 2019, 117, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Engen, J.R.; Beuning, P.J. Escherichia coli Processivity Clamp β from DNA Polymerase III Is Dynamic in Solution. Biochemistry 2011, 50, 5958–5968. [Google Scholar] [CrossRef] [Green Version]
- Purohit, A.; England, J.K.; Douma, L.G.; Tondnevis, F.; Bloom, L.B.; Levitus, M. Electrostatic Interactions at the Dimer Interface Stabilize the E. coli β Sliding Clamp. Biophys. J. 2017, 113, 794–804. [Google Scholar] [CrossRef]
- Perumal, S.K.; Xu, X.; Yan, C.; Ivanov, I.; Benkovic, S.J. Recognition of a Key Anchor Residue by a Conserved Hydrophobic Pocket Ensures Subunit Interface Integrity in DNA Clamps. J. Mol. Biol. 2019, 431, 2493–2510. [Google Scholar] [CrossRef]
- Kelman, Z.; O’Donnell, M. Structural and functional similaritites of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995, 23, 4938. [Google Scholar] [CrossRef] [Green Version]
- Binder, J.K.; Douma, L.G.; Ranjit, S.; Kanno, D.M.; Chakraborty, M.; Bloom, L.B.; Levitus, M. Intrinsic stability and oligomerization dynamics of DNA processivity clamps. Nucleic Acids Res. 2014, 42, 6476–6486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, N.; Turner, J.; Kelman, Z.; Stukenberg, P.T.; Dean, F.; Shechter, D.; Pan, Z.; Hurwitz, J.; O’Donnell, M. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells 1996, 1, 101–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, I.; Chapados, B.R.; McCammon, J.A.; Tainer, J.A. Proliferating cell nuclear antigen loaded onto double-stranded DNA: Dynamics, minor groove interactions and functional implications. Nucleic Acids Res. 2006, 34, 6023–6033. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zheng, F.; O’Donnell, M. Water skating: How polymerase sliding clamps move on DNA. FEBS J. 2021, 288, 7256–7262. [Google Scholar] [CrossRef]
- Smiley, R.D.; Zhuang, Z.; Benkovic, S.J.; Hammes, G.G. Single-Molecule Investigation of the T4 Bacteriophage DNA Polymerase Holoenzyme: Multiple Pathways of Holoenzyme Formation. Biochemistry 2006, 45, 7990–7997. [Google Scholar] [CrossRef] [Green Version]
- Hedglin, M.; Kumar, R.; Benkovic, S.J. Replication Clamps and Clamp Loaders. Cold Spring Harb. Perspect. Biol. 2013, 5, a010165. [Google Scholar] [CrossRef] [Green Version]
- Millar, D.; Trakselis, M.A.; Benkovic, S.J. On the Solution Structure of the T4 Sliding Clamp (gp45). Biochemistry 2004, 43, 12723–12727. [Google Scholar] [CrossRef]
- Jeruzalmi, D.; Yurieva, O.; Zhao, Y.; Young, M.; Stewart, J.; Hingorani, M.; O’Donnell, M.; Kuriyan, J. Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III. Cell 2001, 106, 417–428. [Google Scholar] [CrossRef] [Green Version]
- Podobnik, M.; Weitze, T.F.; O’Donnell, M.; Kuriyan, J. Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit. Structure 2003, 11, 253–263. [Google Scholar] [CrossRef]
- Moolman, M.C.; Krishnan, S.T.; Kerssemakers, J.W.J.; Berg, A.V.D.; Tulinski, P.; Depken, M.; Reyes-Lamothe, R.; Sherratt, D.J.; Dekker, N.H. Slow unloading leads to DNA-bound β2-sliding clamp accumulation in live Escherichia coli cells. Nat. Commun. 2014, 5, 5820. [Google Scholar] [CrossRef] [PubMed]
- Kelch, B.A.; Makino, D.L.; O’Donnell, M.; Kuriyan, J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012, 10, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgescu, R.; Langston, L.; O’Donnell, M. A proposal: Evolution of PCNA’s role as a marker of newly replicated DNA. DNA Repair 2015, 29, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Vivona, J.B.; Kelman, Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 2003, 546, 167–172. [Google Scholar] [CrossRef]
- Geiduschek, E.P.; Kassavetis, G.A. Transcription of the T4 late genes. Virol. J. 2010, 7, 288. [Google Scholar] [CrossRef] [Green Version]
- Blanco, L.; Salas, M. Replication of phage phi 29 DNA with purified terminal protein and DNA polymerase: Synthesis of full-length phi 29 DNA. Proc. Natl. Acad. Sci. USA 1985, 82, 6404–6408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalrymple, B.P.; Kongsuwan, K.; Wijffels, G.; Dixon, N.E.; Jennings, P.A. A universal protein–protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 2001, 98, 11627–11632. [Google Scholar] [CrossRef] [Green Version]
- Warbrick, E. PCNA binding proteins in Drosophila melanogaster: The analysis of a conserved PCNA binding domain. Nucleic Acids Res. 1998, 26, 3925–3932. [Google Scholar] [CrossRef] [Green Version]
- Prestel, A.; Wichmann, N.; Martins, J.M.; Marabini, R.; Kassem, N.; Broendum, S.S.; Otterlei, M.; Nielsen, O.; Willemoës, M.; Ploug, M.; et al. The PCNA interaction motifs revisited: Thinking outside the PIP-box. Cell. Mol. Life Sci. 2019, 76, 4923–4943. [Google Scholar] [CrossRef] [Green Version]
- Camara, J.E.; Skarstad, K.; Crooke, E. Controlled Initiation of Chromosomal Replication in Escherichia coli Requires Functional Hda Protein. J. Bacteriol. 2003, 185, 3244–3248. [Google Scholar] [CrossRef]
- Kim, J.S.; Nanfara, M.T.; Chodavarapu, S.; Jin, K.S.; Babu, V.M.P.; Ghazy, M.A.; Chung, S.; Kaguni, J.M.; Sutton, M.D.; Cho, Y. Dynamic assembly of Hda and the sliding clamp in the regulation of replication licensing. Nucleic Acids Res. 2017, 45, 3888–3905. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kubota, T.; Kurokawa, K.; Crooke, E.; Sekimizu, K. The Initiator Function of DnaA Protein Is Negatively Regulated by the Sliding Clamp of the E. coli Chromosomal Replicase. Cell 1998, 94, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Fernandez, C.; Grosse, K.; Sourjik, V.; Collier, J. The β-sliding clamp directs the localization of HdaA to the replisome in Caulobacter crescentus. Microbiology 2013, 159, 2237–2248. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Saxena, L.; Hoang, N.; Zhang, C.; Lee, L.; Li, W.; Gong, X.; Lu, F.; Sun, H.; Zhang, H. Proliferating cell nuclear antigen interacts with the CRL4 ubiquitin ligase subunit CDT2 in DNA synthesis–induced degradation of CDT1. J. Biol. Chem. 2018, 293, 18879–18889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, A.; Ghose, D.; Thakur, K.; Dutta, D. Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation. PLoS ONE 2018, 13, e0199559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, P.; Tarique, K.F.; Mazumder, M.; Rehman, S.A.A.; Kumari, N.; Gourinath, S. Structural insight into β-Clamp and its interaction with DNA Ligase in Helicobacter pylori. Sci. Rep. 2016, 6, 31181. [Google Scholar] [CrossRef]
- Kukshal, V.; Khanam, T.; Chopra, D.; Singh, N.; Sanyal, S.; Ramachandran, R.M. tuberculosis Sliding β-Clamp Does Not Interact Directly with the NAD+ -Dependent DNA Ligase. PLoS ONE 2012, 7, e35702. [Google Scholar] [CrossRef] [Green Version]
- Beattie, T.R.; Bell, S.D. The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem. Soc. Trans. 2011, 39, 70–76. [Google Scholar] [CrossRef]
- Dovrat, D.; Stodola, J.L.; Burgers, P.M.J.; Aharoni, A. Sequential switching of binding partners on PCNA during in vitro Okazaki fragment maturation. Proc. Natl. Acad. Sci. USA 2014, 111, 14118–14123. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, Y.; Brooks, R.C.; Sverzhinsky, A.; Pascal, J.M.; Tomkinson, A.E. Dynamic DNA-bound PCNA complexes co-ordinate Okazaki fragment synthesis, processing and ligation. J. Mol. Biol. 2020, 432, 166698. [Google Scholar] [CrossRef]
- Sanders, G.M.; Kassavetis, G.A.; Geiduschek, E.P. Dual targets of a transcriptional activator that tracks on DNA. EMBO J. 1997, 16, 3124–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.; Geiduschek, E.; Cascino, A. The role of replication proteins in the regulation of bacteriophage T4 transcription: II. Gene 45 and late transcription uncoupled from replication. J. Mol. Biol. 1975, 96, 539–562. [Google Scholar] [CrossRef]
- Berdis, A.J.; Soumillion, P.; Benkovic, S.J. The carboxyl terminus of the bacteriophage T4 DNA polymerase is required for holoenzyme complex formation. Proc. Natl. Acad. Sci. USA 1996, 93, 12822–12827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Wen, A.; Jin, S.; Gao, B.; Huang, Y.; Feng, Y. Transcription activation by a sliding clamp. Nat. Commun. 2021, 12, 1131. [Google Scholar] [CrossRef]
- Slade, D. Maneuvers on PCNA Rings during DNA Replication and Repair. Genes 2018, 9, 416. [Google Scholar] [CrossRef] [Green Version]
- Pillon, M.C.; Babu, V.M.P.; Randall, J.R.; Cai, J.; Simmons, L.A.; Sutton, M.D.; Guarné, A. The sliding clamp tethers the endonuclease domain of MutL to DNA. Nucleic Acids Res. 2015, 43, 10746–10759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, L.A.; Davies, B.W.; Grossman, A.D.; Walker, G.C. β Clamp Directs Localization of Mismatch Repair in Bacillus subtilis. Mol. Cell 2008, 29, 291–301. [Google Scholar] [CrossRef]
- Hsieh, P.; Zhang, Y. The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2017, 114, 3552–3554. [Google Scholar] [CrossRef] [Green Version]
- Castañeda-García, A.; Prieto, A.I.; Rodríguez-Beltrán, J.; Alonso, N.; Cantillon, D.; Costas, C.; Pérez-Lago, L.; Zegeye, E.D.; Herranz, M.; Plociński, P.; et al. A non-canonical mismatch repair pathway in prokaryotes. Nat. Commun. 2017, 8, 14246. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, N.; Numata, I.; Su’Etsugu, M.; Miyoshi-Akiyama, T. Bacterial EndoMS/NucS acts as a clamp-mediated mismatch endonuclease to prevent asymmetric accumulation of replication errors. Nucleic Acids Res. 2018, 46, 6152–6165. [Google Scholar] [CrossRef]
- Leandro, G.S.; Sykora, P.; Bohr, V.A. The impact of base excision DNA repair in age-related neurodegenerative diseases. Mutat. Res. Mol. Mech. Mutagen. 2015, 776, 31–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lata, K.; Vishwakarma, J.; Kumar, S.; Khanam, T.; Ramachandran, R. Mycobacterium tuberculosis Endonuclease VIII 2 (Nei2) forms a prereplicative BER complex with DnaN: Identification, characterization, and disruption of complex formation. Mol. Microbiol. 2022, 117, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Khanam, T.; Afsar, M.; Shukla, A.; Alam, F.; Kumar, S.; Soyar, H.; Dolma, K.; Pasupuleti, M.; Srivastava, K.K.; Ampapathi, R.S.; et al. M. tuberculosis class II apurinic/apyrimidinic-endonuclease/3′-5′ exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair. Nucleic Acids Res. 2020, 48, 4325–4343. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Moharana, K.; Wallace, S.S.; Doublié, S. Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase. Nucleic Acids Res. 2017, 45, 2897–2909. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Okabe, K.; Hirayama, S.; Chirifu, M.; Ikemizu, S.; Morioka, H.; Nakabeppu, Y.; Yamagata, Y. Structure of the mammalian adenine DNA glycosylase MUTYH: Insights into the base excision repair pathway and cancer. Nucleic Acids Res. 2021, 49, 7154–7163. [Google Scholar] [CrossRef]
- Naiman, K.; Philippin, G.; Fuchs, R.P.; Pagès, V. Chronology in lesion tolerance gives priority to genetic variability. Proc. Natl. Acad. Sci. USA 2014, 111, 5526–5531. [Google Scholar] [CrossRef] [Green Version]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, S.; Fuchs, R.P. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004, 23, 4342–4352. [Google Scholar] [CrossRef] [Green Version]
- Burnouf, D.Y.; Olieric, V.; Wagner, J.; Fujii, S.; Reinbolt, J.; Fuchs, R.P.; Dumas, P. Structural and Biochemical Analysis of Sliding Clamp/Ligand Interactions Suggest a Competition between Replicative and Translesion DNA Polymerases. J. Mol. Biol. 2003, 335, 1187–1197. [Google Scholar] [CrossRef]
- Rêgo, A.T.; Holding, A.N.; Kent, H.; Lamers, M.H. Architecture of the Pol III–clamp–exonuclease complex reveals key roles of the exonuclease subunit in processive DNA synthesis and repair. EMBO J. 2013, 32, 1334–1343. [Google Scholar] [CrossRef]
- Jansen, J.G.; Fousteri, M.I.; de Wind, N. Send in the Clamps: Control of DNA Translesion Synthesis in Eukaryotes. Mol. Cell 2007, 28, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, T.; Wang, Z.; Yi, F.; Li, C.; Guo, W.; Xu, H.; Cui, H.; Dong, X.; Liu, J.; et al. Post-Translational Modifications of PCNA in Control of DNA Synthesis and DNA Damage Tolerance-the Implications in Carcinogenesis. Int. J. Biol. Sci. 2021, 17, 4047–4059. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Katou, Y.; Nakato, R.; Shirahige, K.; Donaldson, A.D. Replication-Coupled PCNA Unloading by the Elg1 Complex Occurs Genome-wide and Requires Okazaki Fragment Ligation. Cell Rep. 2015, 12, 774–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.; Gali, V.K.; Takahashi, T.S.; Kubota, T. PCNA Retention on DNA into G2/M Phase Causes Genome Instability in Cells Lacking Elg1. Cell Rep. 2016, 16, 684–695. [Google Scholar] [CrossRef] [Green Version]
- Shemesh, K.; Sebesta, M.; Pacesa, M.; Sau, S.; Bronstein, A.; Parnas, O.; Liefshitz, B.; Venclovas, C.; Krejci, L.; Kupiec, M. A structure–function analysis of the yeast Elg1 protein reveals the importance of PCNA unloading in genome stability maintenance. Nucleic Acids Res. 2017, 45, 3189–3203. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhuang, Z.; Roccasecca, R.M.; Trakselis, M.A.; Benkovic, S.J. The dynamic processivity of the T4 DNA polymerase during replication. Proc. Natl. Acad. Sci. USA 2004, 101, 8289–8294. [Google Scholar] [CrossRef] [Green Version]
- Indiani, C.; McInerney, P.; Georgescu, R.; Goodman, M.F.; O’Donnell, M. A Sliding-Clamp Toolbelt Binds High- and Low-Fidelity DNA Polymerases Simultaneously. Mol. Cell 2005, 19, 805–815. [Google Scholar] [CrossRef]
- Zhuang, Z.; Johnson, R.E.; Haracska, L.; Prakash, L.; Prakash, S.; Benkovic, S.J. Regulation of polymerase exchange between Polη and Polδ by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 5361–5366. [Google Scholar] [CrossRef] [Green Version]
- Parks, A.R.; Li, Z.; Shi, Q.; Owens, R.M.; Jin, M.M.; Peters, J.E. Transposition into Replicating DNA Occurs through Interaction with the Processivity Factor. Cell 2009, 138, 685–695. [Google Scholar] [CrossRef] [Green Version]
- Gómez, M.J.; Díaz-Maldonado, H.; González-Tortuero, E.; De Saro, F.J.L. Chromosomal Replication Dynamics and Interaction with the β Sliding Clamp Determine Orientation of Bacterial Transposable Elements. Genome Biol. Evol. 2014, 6, 727–740. [Google Scholar] [CrossRef]
- Shibahara, K.-I.; Stillman, B. Replication-Dependent Marking of DNA by PCNA Facilitates CAF-1-Coupled Inheritance of Chromatin. Cell 1999, 96, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Iida, T.; Suetake, I.; Tajima, S.; Morioka, H.; Ohta, S.; Obuse, C.; Tsurimoto, T. PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells 2002, 7, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Young, A.; Autexier, C. PCNA, a focus on replication stress and the alternative lengthening of telomeres pathway. DNA Repair 2021, 100, 103055. [Google Scholar] [CrossRef] [PubMed]
- Hoang, S.M.; O’Sullivan, R.J. Alternative Lengthening of Telomeres: Building Bridges to Connect Chromosome Ends. Trends Cancer 2020, 6, 247–260. [Google Scholar] [CrossRef] [Green Version]
- Witko-Sarsat, V.; Mocek, J.; Bouayad, D.; Tamassia, N.; Ribeil, J.-A.; Candalh, C.; Davezac, N.; Reuter, N.; Mouthon, L.; Hermine, O.; et al. Proliferating cell nuclear antigen acts as a cytoplasmic platform controlling human neutrophil survival. J. Exp. Med. 2010, 207, 2631–2645. [Google Scholar] [CrossRef] [Green Version]
- Olaisen, C.; Kvitvang, H.F.N.; Lee, S.; Almaas, E.; Bruheim, P.; Drabløs, F.; Otterlei, M. The role ofPCNAas a scaffold protein in cellular signaling is functionally conserved between yeast and humans. FEBS Open Bio 2018, 8, 1135–1145. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Venclovas, C. Computational analysis of DNA replicases in double-stranded DNA viruses: Relationship with the genome size. Nucleic Acids Res. 2011, 39, 8291–8305. [Google Scholar] [CrossRef]
- Zuccola, H.; Filman, D.; Coen, D.M.; Hogle, J. The Crystal Structure of an Unusual Processivity Factor, Herpes Simplex Virus UL42, Bound to the C Terminus of Its Cognate Polymerase. Mol. Cell 2000, 5, 267–278. [Google Scholar] [CrossRef]
- Komazin-Meredith, G.; Petrella, R.J.; Santos, W.L.; Filman, D.J.; Hogle, J.M.; Verdine, G.L.; Karplus, M.; Coen, D.M. The Human Cytomegalovirus UL44 C Clamp Wraps around DNA. Structure 2008, 16, 1214–1225. [Google Scholar] [CrossRef] [Green Version]
- Caspari, T.; Dahlen, M.; Kanter-Smoler, G.; Lindsay, H.D.; Hofmann, K.; Papadimitriou, K.; Sunnerhagen, P.; Carr, A.M. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-Related Protein That Associates with Rad1 and Rad9. Mol. Cell. Biol. 2000, 20, 1254–1262. [Google Scholar] [CrossRef]
- Eichinger, C.S.; Jentsch, S. 9-1-1: PCNA’s specialized cousin. Trends Biochem. Sci. 2011, 36, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Kai, M. Role of the Checkpoint Clamp in DNA Damage Response. Biomolecules 2013, 3, 75–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrilla-Castellar, E.R.; Arlander, S.J.; Karnitz, L. Dial 9–1–1 for DNA damage: The Rad9–Hus1–Rad1 (9–1–1) clamp complex. DNA Repair 2004, 3, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Dionne, I.; Nookala, R.K.; Jackson, S.P.; Doherty, A.J.; Bell, S.D. A Heterotrimeric PCNA in the Hyperthermophilic Archaeon Sulfolobus solfataricus. Mol. Cell 2003, 11, 275–282. [Google Scholar] [CrossRef]
- Hamdan, S.M.; Marintcheva, B.; Cook, T.; Lee, S.-J.; Tabor, S.; Richardson, C.C. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. USA 2005, 102, 5096–5101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamdan, S.; Johnson, D.E.; Tanner, N.A.; Lee, J.-B.; Qimron, U.; Tabor, S.; van Oijen, A.; Richardson, C.C. Dynamic DNA Helicase-DNA Polymerase Interactions Assure Processive Replication Fork Movement. Mol. Cell 2007, 27, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Pandey, M.; Inman, J.T.; Yang, Y.; Kashlev, M.; Patel, S.S.; Wang, M.D. T7 replisome directly overcomes DNA damage. Nat. Commun. 2015, 6, 10260. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.E.; Takahashi, M.; Hamdan, S.M.; Lee, S.-J.; Richardson, C.C. Exchange of DNA polymerases at the replication fork of bacteriophage T7. Proc. Natl. Acad. Sci. USA 2007, 104, 5312–5317. [Google Scholar] [CrossRef] [Green Version]
- Loparo, J.J.; Kulczyk, A.W.; Richardson, C.C.; van Oijen, A.M. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc. Natl. Acad. Sci. USA 2011, 108, 3584–3589. [Google Scholar] [CrossRef] [Green Version]
- Altieri, A.S.; Kelman, Z. DNA Sliding Clamps as Therapeutic Targets. Front. Mol. Biosci. 2018, 5, 87. [Google Scholar] [CrossRef]
- André, C.; Martiel, I.; Wolff, P.; Landolfo, M.; Lorber, B.; Da Veiga, C.S.; Dejaegere, A.; Dumas, P.; Guichard, G.; Olieric, V.; et al. Interaction of a Model Peptide on Gram Negative and Gram Positive Bacterial Sliding Clamps. ACS Infect. Dis. 2019, 5, 1022–1034. [Google Scholar] [CrossRef] [PubMed]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A Bacterial Toxin Inhibits DNA Replication Elongation through a Direct Interaction with the β Sliding Clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Nedal, A.; Ræder, S.B.; Dalhus, B.; Helgesen, E.; Forstrøm, R.J.; Lindland, K.; Sumabe, B.K.; Martinsen, J.H.; Kragelund, B.B.; Skarstad, K.; et al. Peptides containing the PCNA interacting motif APIM bind to the β-clamp and inhibit bacterial growth and mutagenesis. Nucleic Acids Res. 2020, 48, 5540–5554. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Whittell, L.R.; Wang, Y.; Jergic, S.; Liu, M.; Harry, E.; Dixon, N.; Beck, J.; Kelso, M.; Oakley, A.J. Discovery of Lead Compounds Targeting the Bacterial Sliding Clamp Using a Fragment-Based Approach. J. Med. Chem. 2014, 57, 2799–2806. [Google Scholar] [CrossRef] [PubMed]
- Kling, A.; Lukat, P.; Almeida, D.V.; Bauer, A.; Fontaine, E.; Sordello, S.; Zaburannyi, N.; Herrmann, J.; Wenzel, S.C.; König, C.; et al. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 2015, 348, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-C. PCNA: A silent housekeeper or a potential therapeutic target? Trends Pharmacol. Sci. 2014, 35, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Wortman, M.; Dillehay, K.L.; Seibel, W.L.; Evelyn, C.R.; Smith, S.J.; Malkas, L.H.; Zheng, Y.; Lu, S.; Dong, Z. Small-Molecule Targeting of Proliferating Cell Nuclear Antigen Chromatin Association Inhibits Tumor Cell Growth. Mol. Pharmacol. 2012, 81, 811–819, Erratum in Mol. Pharmacol. 2013, 83, 719. [Google Scholar] [CrossRef]




| Process | Proteins Interacting with Eukaryotic PCNA | Proteins Interacting with Bacterial Sliding Clamp |
|---|---|---|
| DNA replication | DNA Polymerase δ, DNA Polymerase ε, Replication factor (RfC1, Rfc3, Rfc4), Flap endonuclease (FEN-1), DNA ligase 1, DNA topoisomerase 1, Cdt1, Topo Iiα | DNA Polymerase III, DnaA, Clamp loader complex, DNA Polymerase I, DNA ligase |
| DNA repair | DNA mismatch repair protein Msh3, Msh6, Mlh1, Exonuclease 1, UNG2, MPG, PARP-1, APE1, APE2, XRCC1, WRN, BLM, RECQ5, NTH1, hMYH, XPG | MutL, MutS, NucS, Nei2, XthA, DNA Polymerase IV, DNA Polymerase V |
| DNA damage | DNA Polymerase η, DNA Polymerase ζ, DNA Polymerase λ, DNA Polymerase κ, DNA repair protein REV1, E3 ubiquitin-protein ligase RAD18 and Rad5 | |
| Chromatin Assembly | WSTF, DNMT1, HDAC1, p300, CAF-1 | _ |
| Cell cycle control | p21 (C1P1/WAF1), P27 (KIP2), Cyclin D1, MCL1, P15 (PAF), CDK2, ING1b, Gadd45, MyD118, CR6, P53, MDM2 | DnaA, Hda |
| Sister chromatid cohesion | ESCO1, ESCO2, Chl1, Ctf18 | _ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulye, M.; Singh, M.I.; Jain, V. From Processivity to Genome Maintenance: The Many Roles of Sliding Clamps. Genes 2022, 13, 2058. https://doi.org/10.3390/genes13112058
Mulye M, Singh MI, Jain V. From Processivity to Genome Maintenance: The Many Roles of Sliding Clamps. Genes. 2022; 13(11):2058. https://doi.org/10.3390/genes13112058
Chicago/Turabian StyleMulye, Meenakshi, Manika Indrajit Singh, and Vikas Jain. 2022. "From Processivity to Genome Maintenance: The Many Roles of Sliding Clamps" Genes 13, no. 11: 2058. https://doi.org/10.3390/genes13112058
APA StyleMulye, M., Singh, M. I., & Jain, V. (2022). From Processivity to Genome Maintenance: The Many Roles of Sliding Clamps. Genes, 13(11), 2058. https://doi.org/10.3390/genes13112058




