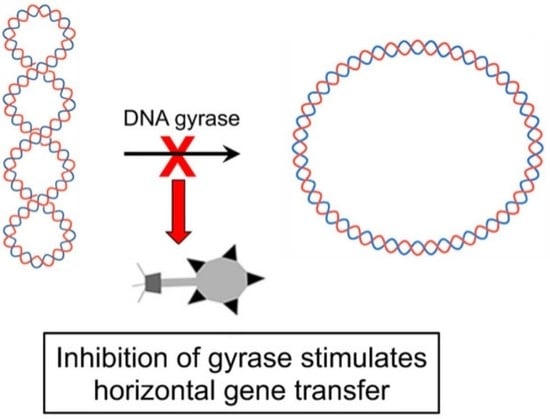
DNA Gyrase Inhibitors Increase the Frequency of Bacteriophage-like RcGTA-Mediated Gene Transfer in Rhodobacter capsulatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Recombinant DNA Techniques, Plasmids, and Primers
2.3. Transduction Assay
2.4. RcGTA Bioassay
2.5. SDS-PAGE and Western Blot
2.6. Disruption of RcGTA Gene g4
2.7. Conjugation
2.8. Construction of gyrB Overexpression Plasmid
2.9. Statistical Analysis
3. Results
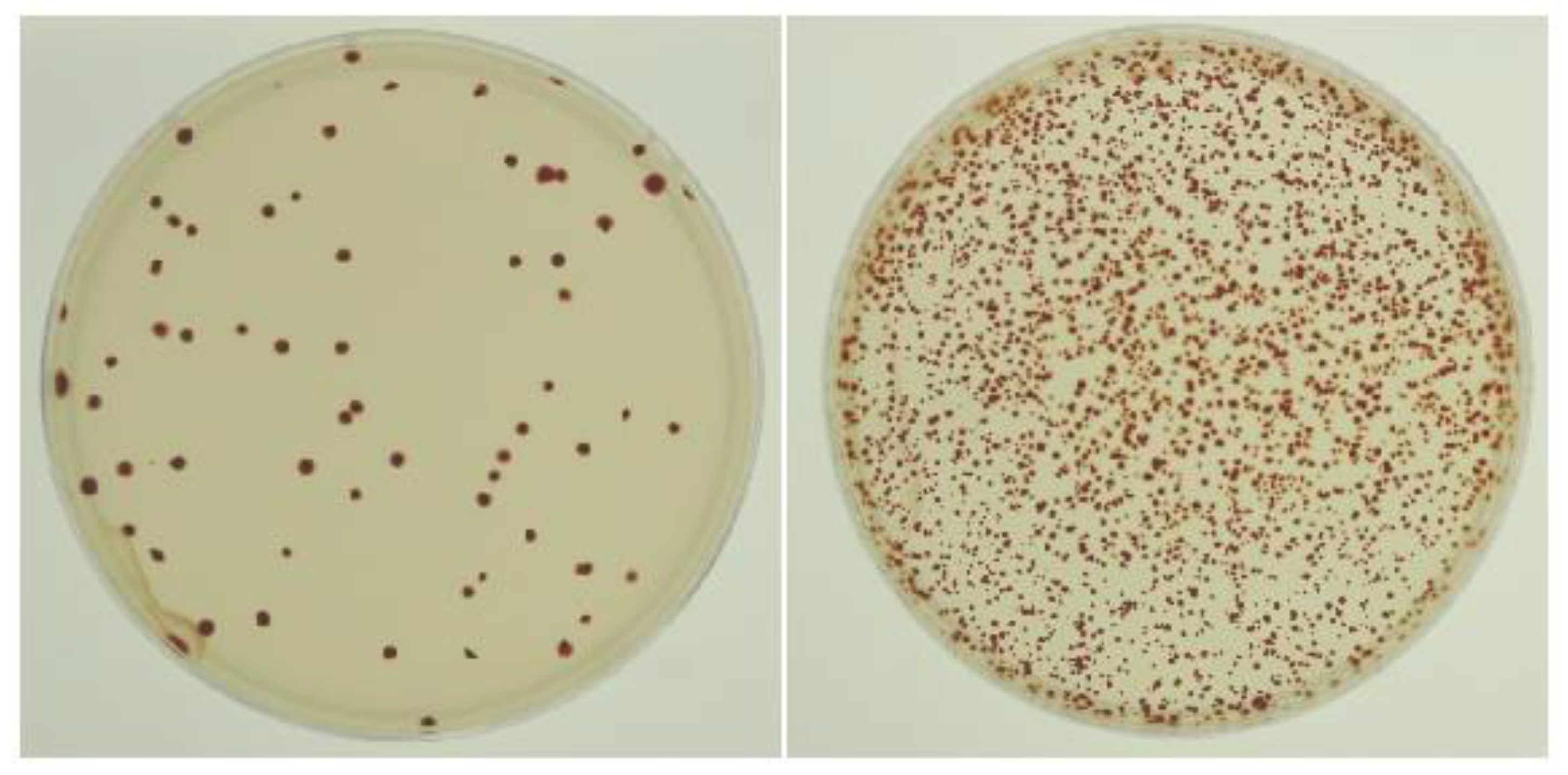
3.1. Development of the Transduction Assay
3.2. DNA Gyrase Inhibitors Increase the Frequency of RcGTA-Mediated Gene Transfer
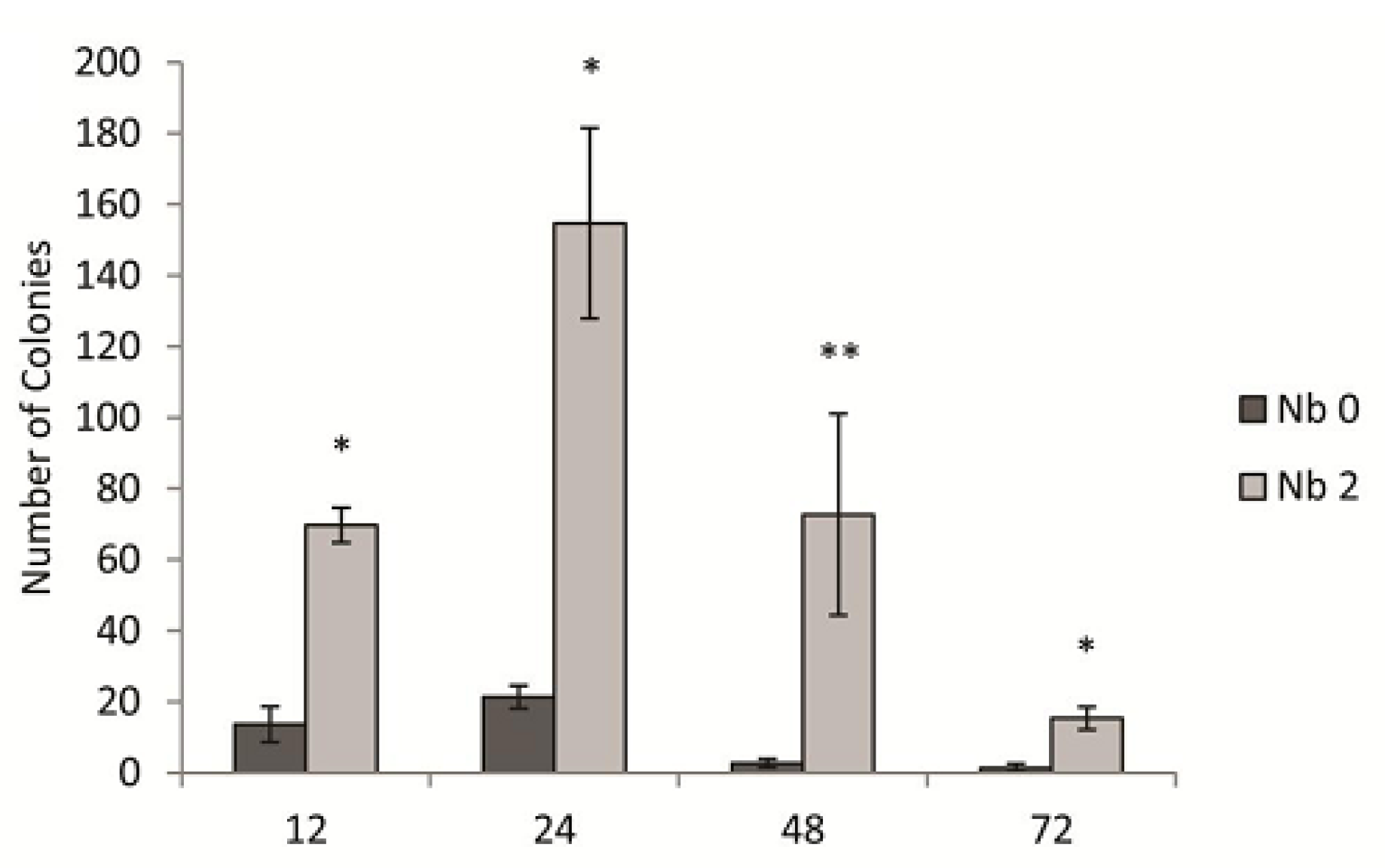
3.3. Kinetics of the Novobiocin Effect in the Transduction Assay
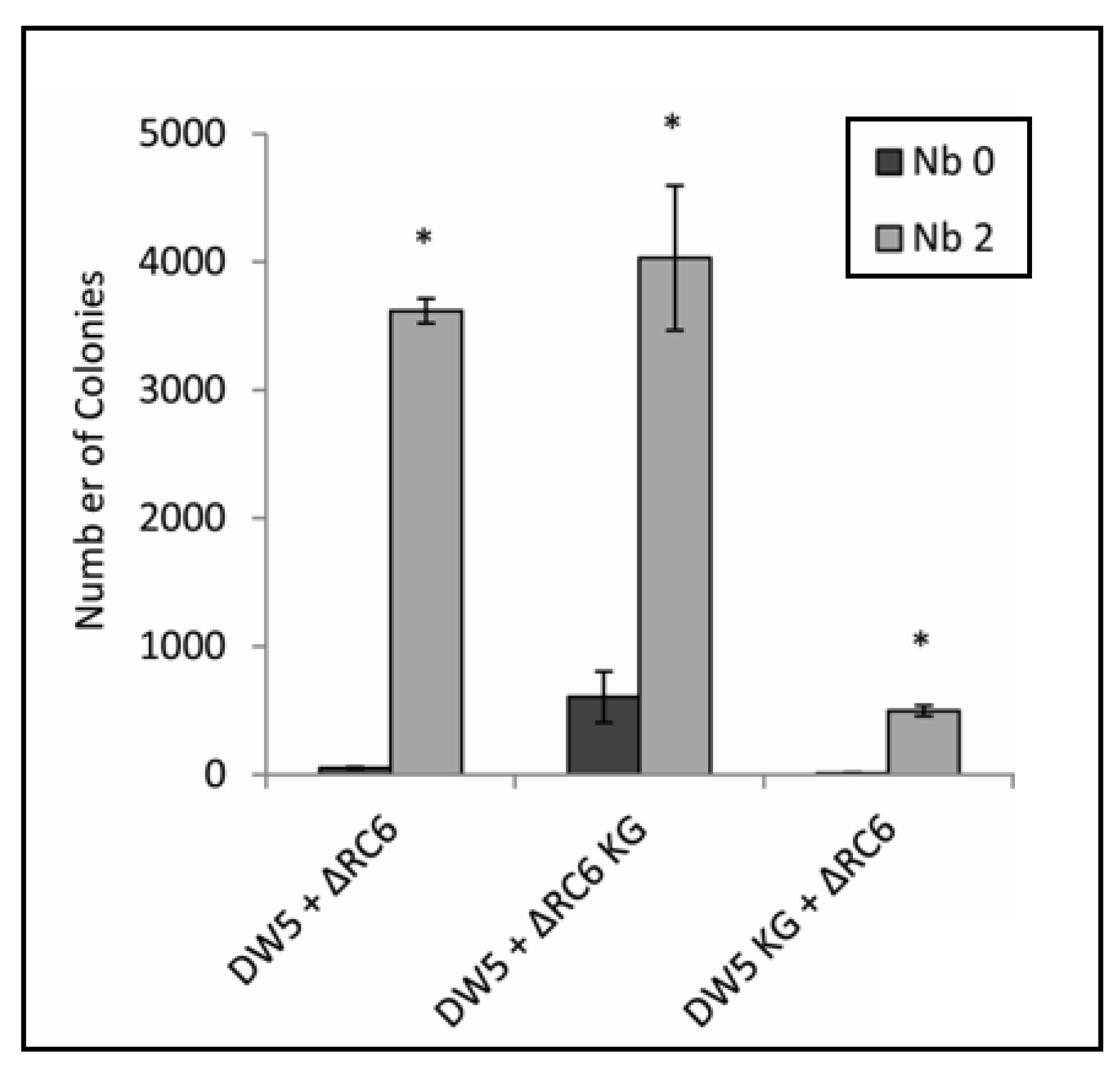
3.4. Overexpression of gyrB Increases Resistance to Novobiocin and Inhibits the Novobiocin-Dependent Increase in Gene Transfer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kogay, R.; Koppenhöfer, S.; Beatty, J.T.; Kuhn, J.H.; Lang, A.S.; Zhaxybayeva, O. Formal recognition and classification of gene transfer agents as viriforms. Virus Evol. in press. [CrossRef]
- Lang, A.S.; Zhaxybayeva, O.; Beatty, J.T. Gene transfer agents: Phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012, 10, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Bardy, P.; Fuzik, T.; Hrebik, D.; Pantucek, R.; Thomas Beatty, J.; Plevka, P. Structure and mechanism of DNA delivery of a gene transfer agent. Nat. Commun. 2020, 11, 3034. [Google Scholar] [CrossRef] [PubMed]
- Westbye, A.B.; Leung, M.M.; Florizone, S.; Taylor, T.A.; Johnson, J.A.; Fogg, P.C.; Beatty, J.T. Phosphate concentration and the putative sensor-kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent (RcGTA). J. Bacteriol. 2013, 195, 5025–5040. [Google Scholar] [CrossRef]
- Hynes, A.P.; Mercer, R.G.; Watton, D.E.; Buckley, C.B.; Lang, A.S. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol. Microbiol. 2012, 85, 314–325. [Google Scholar] [CrossRef]
- Westbye, A.B.; Kuchinski, K.; Yip, C.K.; Beatty, J.T. The gene transfer agent RcGTA contains head spikes needed for binding to the Rhodobacter capsulatus polysaccharide cell capsule. J. Mol. Biol. 2016, 428, 477–491. [Google Scholar] [CrossRef]
- Fogg, P.C.; Westbye, A.B.; Beatty, J.T. One for all or all for one: Heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS ONE 2012, 7, e43772. [Google Scholar] [CrossRef]
- Ding, H.; Grull, M.P.; Mulligan, M.E.; Lang, A.S.; Beatty, J.T. Induction of Rhodobacter capsulatus gene transfer agent gene expression is a bistable stochastic process repressed by an extracellular calcium-binding RTX protein homologue. J. Bacteriol. 2019, 201, e00430-19. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Ding, H.; Johnson, J.A.; Beatty, J.T. Homologues of genetic transformation DNA import genes are required for Rhodobacter capsulatus gene transfer agent recipient capability regulated by the response regulator CtrA. J. Bacteriol. 2015, 197, 2653–2663. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Stevens, A.; Jun, D.; Mercer, R.; Lang, A.S.; Beatty, J.T. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol. Microbiol. 2013, 87, 802–817. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Ding, H.; Beatty, J.T. Rhodobacter capsulatus DprA is essential for RecA-mediated gene transfer agent (RcGTA) recipient capability regulated by quorum-sensing and the CtrA response regulator. Mol. Microbiol. 2014, 92, 1260–1278. [Google Scholar] [CrossRef]
- Westbye, A.B.; Beatty, J.T.; Lang, A.S. Guaranteeing a captive audience: Coordinated regulation of gene transfer agent (GTA) production and recipient capability by cellular regulators. Curr. Opin. Microbiol. 2017, 38, 122–129. [Google Scholar] [CrossRef]
- Goh, E.B.; Yim, G.; Tsui, W.; McClure, J.; Surette, M.G.; Davies, J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 2002, 99, 17025–17030. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, X.; Kay, E.; Schneider, D.; Shuman, H.A. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J. Bacteriol. 2011, 193, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Beaber, J.W.; Hochhut, B.; Waldor, M.K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004, 427, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; McDaniel, A.D.; Wolf, L.E.; Keusch, G.T.; Waldor, M.K.; Acheson, D.W. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 2000, 181, 664–670. [Google Scholar] [CrossRef]
- Stanton, T.B.; Humphrey, S.B.; Sharma, V.K.; Zuerner, R.L. Collateral effects of antibiotics: Carbadox and metronidazole induce VSH-1 and facilitate gene transfer among Brachyspira hyodysenteriae strains. Appl. Environ. Microbiol. 2008, 74, 2950–2956. [Google Scholar] [CrossRef]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA Topoisomerases. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef]
- Heide, L. New aminocoumarin antibiotics as gyrase inhibitors. Int. J. Med. Microbiol. 2014, 304, 31–36. [Google Scholar] [CrossRef]
- Mustaev, A.; Malik, M.; Zhao, X.; Kurepina, N.; Luan, G.; Oppegard, L.M.; Hiasa, H.; Marks, K.R.; Kerns, R.J.; Berger, J.M.; et al. Fluoroquinolone-gyrase-DNA complexes: Two modes of drug binding. J. Biol. Chem. 2014, 289, 12300–12312. [Google Scholar] [CrossRef]
- Blower, T.R.; Williamson, B.H.; Kerns, R.J.; Berger, J.M. Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2016, 113, 1706–1713. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Beatty, J.T.; Gest, H. Biosynthetic and bioenergetic functions of citric acid cycle reactions in Rhodopseudomonas capsulata. J. Bacteriol. 1981, 148, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.D.; Weaver, P.F.; Gest, H. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch. Microbiol. 1975, 105, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Marrs, B. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 1974, 71, 971–973. [Google Scholar] [CrossRef]
- Yen, H.C.; Marrs, B. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 1976, 126, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Beatty, J.T.; Cohen, S.N.; Belasco, J.G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell 1988, 52, 609–619. [Google Scholar] [CrossRef]
- Wong, D.K.; Collins, W.J.; Harmer, A.; Lilburn, T.G.; Beatty, J.T. Directed mutagenesis of the Rhodobacter capsulatus puhA gene and orf 214: Pleiotropic effects on photosynthetic reaction center and light-harvesting 1 complexes. J. Bacteriol. 1996, 178, 2334–2342. [Google Scholar] [CrossRef]
- LeBlanc, H.N.; Beatty, J.T. Rhodobacter capsulatus puc operon: Promoter location, transcript sizes and effects of deletions on photosynthetic growth. J. Gen. Microbiol. 1993, 139, 101–109. [Google Scholar] [CrossRef][Green Version]
- Leung, M.M.; Brimacombe, C.A.; Spiegelman, G.B.; Beatty, J.T. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol. Microbiol. 2012, 83, 759–774. [Google Scholar] [CrossRef]
- Taylor, T.A. Evolution and Regulation of the Gene Transfer Agent (GTA) of Rhodobacter capsulatus; University of British Columbia: Vancouver, BC, Canada, 2004. [Google Scholar]
- Fellay, R.; Frey, J.; Krisch, H. Interposon mutagenesis of soil and water bacteria: A family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 1987, 52, 147–154. [Google Scholar] [CrossRef]
- Lang, A.S.; Beatty, J.T. Genetic analysis of a bacterial genetic exchange element: The gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA 2000, 97, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.P.; Cohen, S.N.; Clark, W.G.; Marrs, B.L. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J. Bacteriol. 1983, 154, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Barany, F. Single-stranded hexameric linkers: A system for in-phase insertion mutagenesis and protein engineering. Gene 1985, 37, 111–123. [Google Scholar] [CrossRef]
- Katzke, N.; Arvani, S.; Bergmann, R.; Circolone, F.; Markert, A.; Svensson, V.; Jaeger, K.-E.; Heck, A.; Drepper, T. A novel T7 RNA polymerase dependent expression system for high-level protein production in the phototrophic bacterium Rhodobacter capsulatus. Protein Expr. Purif. 2010, 69, 137–146. [Google Scholar] [CrossRef]
- Solioz, M.; Yen, H.C.; Marris, B. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J. Bacteriol. 1975, 123, 651–657. [Google Scholar] [CrossRef]
- Yen, H.C.; Hu, N.T.; Marrs, B.L. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 1979, 131, 157–168. [Google Scholar] [CrossRef]
- Bearson, B.L.; Brunelle, B.W. Fluoroquinolone induction of phage-mediated gene transfer in multidrug-resistant Salmonella. Int. J. Antimicrob. Agents 2015, 46, 201–204. [Google Scholar] [CrossRef]
- Bearson, B.L.; Allen, H.K.; Brunelle, B.W.; Lee, I.S.; Casjens, S.R.; Stanton, T.B. The agricultural antibiotic carbadox induces phage-mediated gene transfer in Salmonella. Front. Microbiol. 2014, 5, 52. [Google Scholar] [CrossRef]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef]
- Kowalczykowski, S.C.; Dixon, D.A.; Eggleston, A.K.; Lauder, S.D.; Rehrauer, W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994, 58, 401–465. [Google Scholar] [CrossRef]
- Dorman, C.J.; Dorman, M.J. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys. Rev. 2016, 8, 209–220. [Google Scholar] [CrossRef]
- Peter, B.J.; Arsuaga, J.; Breier, A.M.; Khodursky, A.B.; Brown, P.O.; Cozzarelli, N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004, 5, R87. [Google Scholar] [CrossRef] [PubMed]
- Kummerer, K. Resistance in the environment. J. Antimicrob. Chemother. 2004, 54, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Tomasch, J.; Wang, H.; Hall, A.T.K.; Patzelt, D.; Preusse, M.; Petersen, J.; Brinkmann, H.; Bunk, B.; Bhuju, S.; Jarek, M.; et al. Packaging of Dinoroseobacter shibae DNA into gene transfer agent particles is not random. Genome Biol. Evol. 2018, 10, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Nagao, N.; Yamamoto, J.; Komatsu, H.; Suzuki, H.; Hirose, Y.; Umekage, S.; Ohyama, T.; Kikuchi, Y. The gene transfer agent-like particle of the marine phototrophic bacterium Rhodovulum sulfidophilum. Biochem. Biophys. Rep. 2015, 4, 369–374. [Google Scholar] [CrossRef]
- Biers, E.J.; Wang, K.; Pennington, C.; Belas, R.; Chen, F.; Moran, M.A. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl. Environ. Microbiol. 2008, 74, 2933–2939. [Google Scholar] [CrossRef] [PubMed]
- Kogay, R.; Neely, T.B.; Birnbaum, D.P.; Hankel, C.R.; Shakya, M.; Zhaxybayeva, O. Machine-learning classification suggests that many alphaproteobacterial prophages may instead be gene transfer agents. Genome Biol. Evol. 2019, 11, 2941–2953. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.S.; Beatty, J.T. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 2007, 15, 54–62. [Google Scholar] [CrossRef]
- Power, J.J.; Pinheiro, F.; Pompei, S.; Kovacova, V.; Yuksel, M.; Rathmann, I.; Forster, M.; Lassig, M.; Maier, B. Adaptive evolution of hybrid bacteria by horizontal gene transfer. Proc. Natl. Acad. Sci. USA 2021, 118, e2007873118. [Google Scholar] [CrossRef]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]


| Strain | Source | Description |
|---|---|---|
| B10 | [25] | wild type |
| SB1003 | [26] | RifR B10 derivative |
| ΔRC6 | [27] | B10 background, puf operon knockout, KanR |
| DW5 | [28] | SB1003 background, translationally in-frame puhA deletion |
| SBΔRC6 | this study | transduction of ΔRC6 puf operon knockout into SB1003, KanR |
| ΔRC6_Δg4 | this study | ΔRC6 with g4 (GTA protease gene) disruption, KanR/SpcR |
| DW5_Δg4 | this study | DW5 with g4 (GTA protease gene) disruption, KanR |
| ΔLHII | [29] | SBI003 background, puc operon knockout, SpcR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernelot-Moens, R.; Beatty, J.T. DNA Gyrase Inhibitors Increase the Frequency of Bacteriophage-like RcGTA-Mediated Gene Transfer in Rhodobacter capsulatus. Genes 2022, 13, 2071. https://doi.org/10.3390/genes13112071
Bernelot-Moens R, Beatty JT. DNA Gyrase Inhibitors Increase the Frequency of Bacteriophage-like RcGTA-Mediated Gene Transfer in Rhodobacter capsulatus. Genes. 2022; 13(11):2071. https://doi.org/10.3390/genes13112071
Chicago/Turabian StyleBernelot-Moens, Rachel, and J. Thomas Beatty. 2022. "DNA Gyrase Inhibitors Increase the Frequency of Bacteriophage-like RcGTA-Mediated Gene Transfer in Rhodobacter capsulatus" Genes 13, no. 11: 2071. https://doi.org/10.3390/genes13112071
APA StyleBernelot-Moens, R., & Beatty, J. T. (2022). DNA Gyrase Inhibitors Increase the Frequency of Bacteriophage-like RcGTA-Mediated Gene Transfer in Rhodobacter capsulatus. Genes, 13(11), 2071. https://doi.org/10.3390/genes13112071