A Missense Mutation (c.1037 G > C, p. R346P) in PAPSS2 Gene Results in Autosomal Recessive form of Brachyolmia Type 1 (Hobaek Form) in A Consanguineous Family
Abstract
1. Introduction
2. Material and Method
2.1. Recruitment of Family and Collection of Blood Samples
2.2. Whole Exome Sequencing
2.3. PCR
2.4. Sanger Sequencing
2.5. Cloning of PAPSS2 Gene
2.6. Cell Culture, Transfection and Immunoblotting
2.7. Multiple Sequence Alignment Analysis
2.8. Protein Model Building
3. Results
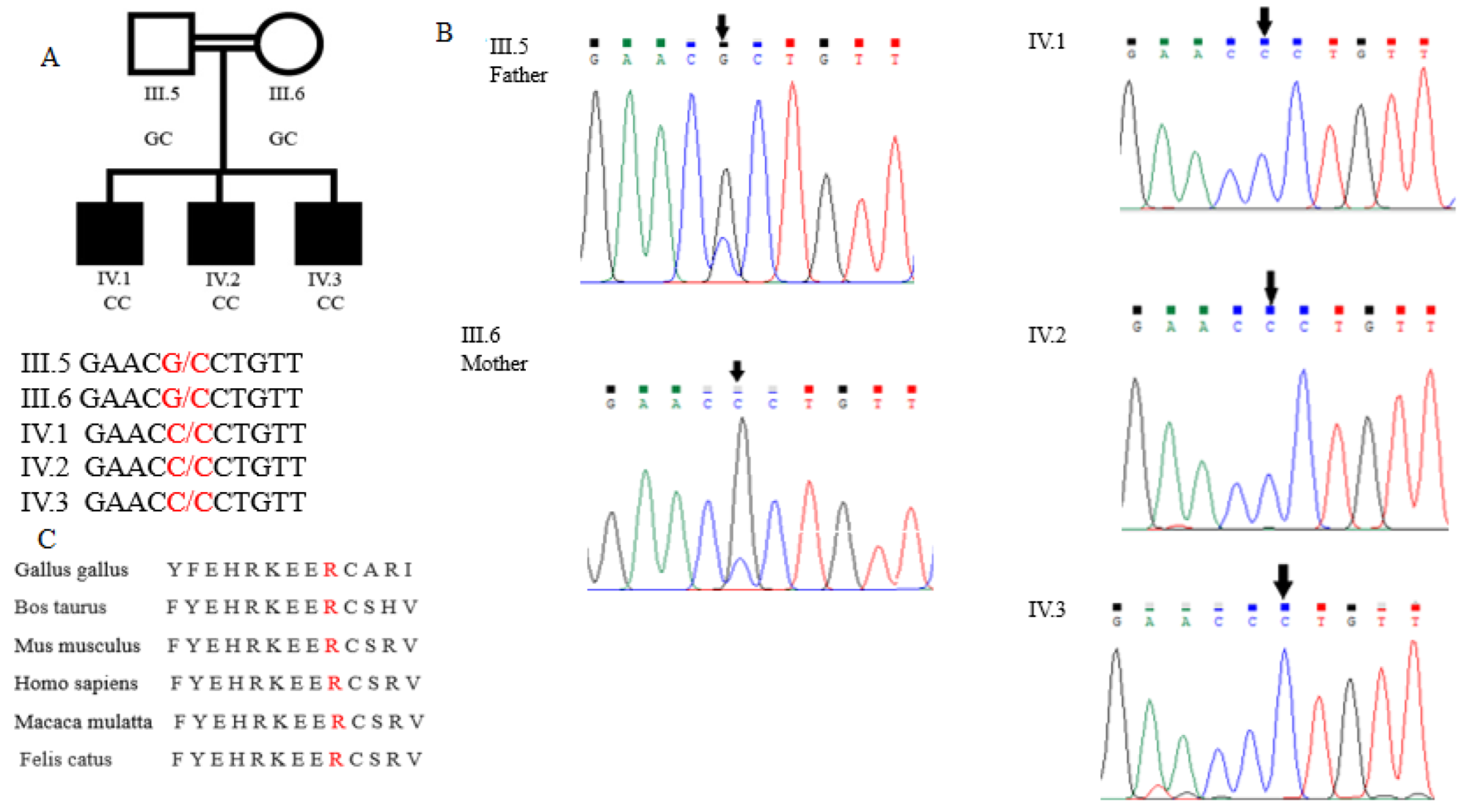
3.1. Phenotype and Radiographic Findings
3.2. PAPSS2 Gene Mutation
3.3. Full-Length and Mutated PAPSS2 Gene Expression
3.4. PAPSS2 Protein Structure Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoo, J.J.; Oliphant, M. Two sibs with brachyolmia type Hobaek: Five year follow-up through puberty. Am. J. Med. Genet. 2002, 116A, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Miyake, N.; Elcioglu, N.H.; Iida, A.; Isguven, P.; Dai, J.; Murakami, N.; Takamura, K.; Cho, T.-J.; Kim, O.-H.; Hasegawa, T.; et al. PAPSS2 mutations cause autosomal recessive brachyolmia. J. Med. Genet. 2012, 49, 533–538. [Google Scholar] [CrossRef]
- Eltana, M.; Abali, Z.Y.; Ates, E.A.; Kirkgoz, T.; Kaygusuz, S.B.; Türkyılmaz, A.; Bereket, A.; Turan, S.; Guran, T. Low DHEAS Concentration in a Girl Presenting with Short Stature and Premature Pubarche: A Novel PAPSS2 Gene Mutation. Horm. Res. Paediatr. 2019, 92, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Shohat, M.; Lachman, R.; Gruber, H.E.; Rimoin, D.L. Brachyolmia: Radiographic and genetic evidence of heterogeneity. Am. J. Med. Genet. 1989, 33, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Rock, M.J.; Prenen, J.; Funari, V.A.; Funari, T.L.; Merriman, B.; Nelson, S.F.; Lachman, R.S.; Wilcox, W.R.; Reyno, S.; Quadrelli, R.; et al. Gain-of-function mutations in TRPV4 cause autosomal dominant brachyolmia. Nat. Genet. 2008, 40, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.; Wang, L.L.; Li, Y.; Zhou, S.F. Structure, function and polymorphism of human cytosolic sulfotransferases. Curr. Drug Metabol. 2008, 9, 99–105. [Google Scholar]
- Mizumoto, S.; Ikegawa, S.; Sugahara, K. Human genetic disorders caused by mutations in the genes encoding biosynthetic enzymes for sulfated glycosaminoglycans. J. Biol. Chem. 2013, 288, 10953–10961. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Guo, F.; Ning, Y.; Zhi, X.; Wang, X.; Chen, S.; Yin, L.; Li, X. Effect of estrogen sulfation by SULT1E1 and PAPSS on the development of estrogen-dependent cancers. Cancer Sci. 2012, 103, 1000–1009. [Google Scholar] [CrossRef]
- Falany, C.N. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997, 11, 206–216. [Google Scholar] [CrossRef]
- Mustafa, S.; Bukhari, F.; Aftab, M.N.; Asif, M.; Amjad, M.; Ijaz, M.; Latif, M.; Iqbal, F. A missense mutation in COL10A1 gene causing Metaphyseal chondrodysplasia, Schmid type in a Pakistani consanguineous family. Pak. J. Zool. 2022, 54, 1447–1450. [Google Scholar] [CrossRef]
- Yasin, S.; Mustafa, S.; Ayesha, A.; Latif, M.; Hassan, M.; Faisal, M.; Makitie, O.; Iqbal, F.; Naz, S. A novel homozygous missense variant in MATN3 causes Spondylo-epimetaphyseal dysplasia Matrilin 3 type in a consanguineous family. Eur. J. Med. Genet. 2020, 63, 103958. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, S.; Akhtar, Z.; Latif, M.; Hassan, M.; Faisal, M.; Iqbal, F. A novel nonsense mutation in NPR2 gene causing Acromesomelic dysplasia, type Maroteaux in a consanguineous family in Southern Punjab (Pakistan). Genes Genom. 2020, 42, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yang, D.; Omerallo, A.K.; Zavialov, A.V.; Toro, C.; Zavialov, A.V.; Stone, D.L.; Chae, J.J.; Rosenzweig, S.D.; Bishop, K. Early-onset stroke and vasculopathy associated with mutations in ADA2. N. Eng. J. Med. 2014, 370, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Iida, A.; Simsek-Kiper, P.O.; Mizumoto, S.; Hoshino, T.; Elcioglu, N.; Horemuzova, E.; Geiberger, S.; Yesil, G.; Kayserili, H.; Utine, G.E.; et al. Clinical and radiographic features of the autosomal recessive form of brachyolmia caused by PAPSS2 mutations. Hum. Mutat. 2013, 34, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.H.; Otterness, D.M.; Freimuth, R.R.; Carlini, E.; Wood, T.C.; Mitchell, S.; Moon, E.; Kim, U.; Xu, J.; Siciliano, M.; et al. Human 3-phosphoadenosine 5-phosphosulfate synthetase 1 (PAPSS1) and PAPSS2: Gene cloning, characterization and chromosomal localization. Biochem. Biophy. Res. Commun. 2000, 268, 437–444. [Google Scholar] [CrossRef]
- Stelzer, C.; Brimmer, A.; Hermanns, P.; Zabel, B.; Dietz, U.H. Expression prolie of Papss2 (3phosphoadenosine5-phosphosulfate synthase 2) during cartilage formation and skeletal development in the mouse embryo. Dev. Dyn. 2017, 236, 1313–1318. [Google Scholar] [CrossRef]
- Brylski, O.; Shrestha, P.; House, P.J.; Gnutt, P.; Mueller, J.W.; Ebbinghaus, S. Disease-Related Protein Variants of the Highly Conserved Enzyme PAPSS2 Show Marginal Stability and Aggregation in Cells. Front. Mol. Biosci. 2022, 89, 860387. [Google Scholar] [CrossRef]
- Bownass, L.; Abbs, S.; Armstrong, R.; Baujat, G.; Behzadi, G.; Berentsen, R.D.; Burren, C.; Calder, A.; Cormier-Daire, V.; Newbury-Ecob, R.; et al. PAPSS2-related brachyolmia: Clinical and radiological phenotype in 18 new cases. Am. J. Med. Genet. 2019, 179, 1884–1894. [Google Scholar] [CrossRef]
- Melissa Perez-Garcia, E.; Whalen, P.; Gurtunca, N. Novel Inactivating Homozygous PAPSS2 Mutation in Two Siblings With Disproportionate Short Stature. AACE Clin. Case Rep. 2022, 8, 89–92. [Google Scholar] [CrossRef]
- Tüysüz, B.; Yılmaz, S.; Gül, E.; Kolb, L.; Bilguvar, K.; Evliyaoğlu, O.; Günel, M. Spondyloepimetaphyseal dysplasia Pakistani type: Expansion of the phenotype. Am. J. Med. Genet A 2013, 161, 1300–1308. [Google Scholar] [CrossRef]
- Noordam, C.; Dhir, V.; McNelis, J.C.; Schlereth, F.; Hanley, N.A.; Krone, N.; Smeitink, J.A.; Smeets, R.; Sweep, F.C.; der Grinten, H.L.C.-V.; et al. Inactivating PAPSS2 mutations in a patient with premature pubarche. N. Eng. J. Med. 2009, 360, 2310–2318. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustafa, S.; Hussain, M.F.; Latif, M.; Ijaz, M.; Asif, M.; Hassan, M.; Faisal, M.; Iqbal, F. A Missense Mutation (c.1037 G > C, p. R346P) in PAPSS2 Gene Results in Autosomal Recessive form of Brachyolmia Type 1 (Hobaek Form) in A Consanguineous Family. Genes 2022, 13, 2096. https://doi.org/10.3390/genes13112096
Mustafa S, Hussain MF, Latif M, Ijaz M, Asif M, Hassan M, Faisal M, Iqbal F. A Missense Mutation (c.1037 G > C, p. R346P) in PAPSS2 Gene Results in Autosomal Recessive form of Brachyolmia Type 1 (Hobaek Form) in A Consanguineous Family. Genes. 2022; 13(11):2096. https://doi.org/10.3390/genes13112096
Chicago/Turabian StyleMustafa, Saima, Malik Fiaz Hussain, Muhammad Latif, Maryam Ijaz, Muhammad Asif, Mubashir Hassan, Muhammad Faisal, and Furhan Iqbal. 2022. "A Missense Mutation (c.1037 G > C, p. R346P) in PAPSS2 Gene Results in Autosomal Recessive form of Brachyolmia Type 1 (Hobaek Form) in A Consanguineous Family" Genes 13, no. 11: 2096. https://doi.org/10.3390/genes13112096
APA StyleMustafa, S., Hussain, M. F., Latif, M., Ijaz, M., Asif, M., Hassan, M., Faisal, M., & Iqbal, F. (2022). A Missense Mutation (c.1037 G > C, p. R346P) in PAPSS2 Gene Results in Autosomal Recessive form of Brachyolmia Type 1 (Hobaek Form) in A Consanguineous Family. Genes, 13(11), 2096. https://doi.org/10.3390/genes13112096






