Identification of Key Genes and Pathways Associated with Preeclampsia by a WGCNA and an Evolutionary Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray Data
2.2. Differentially Expressed Gene Screening
2.3. WGCNA
2.4. Gene Annotation and Enrichment Analysis
2.5. Construction of a Human Protein–Protein Interaction (PPI) Network
2.6. PPI Subnetworks among Proteins Encoded by PE-Associated Genes
2.7. Enrichment Analysis of HAR and PS Genes
3. Results
3.1. WGCNA Results
3.2. GO Analysis
3.3. PPI Analysis and the Identification of Densely Connected Clusters
3.4. Enrichment of HAR and PS Genes in the PPI Clusters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duley, L. The global impact of pre-eclampsia and eclampsia. Semin. Perinatol. 2009, 33, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Steegers, E.A.; von Dadelszen, P.; Duvekot, J.J.; Pijnenborg, R. Pre-eclampsia. Lancet 2010, 376, 631–644. [Google Scholar] [CrossRef]
- Abalos, E.; Cuesta, C.; Grosso, A.L.; Chou, D.; Say, L. Global and regional estimates of preeclampsia and eclampsia: A systematic review. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 170, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Phipps, E.A.; Thadhani, R.; Benzing, T.; Karumanchi, S.A. Pre-eclampsia: Pathogenesis, novel diagnostics and therapies. Nat. Rev. Nephrol. 2019, 15, 275–289. [Google Scholar] [CrossRef]
- Stekkinger, E.; Zandstra, M.; Peeters, L.L.H.; Spaanderman, M.E.A. Early-onset preeclampsia and the prevalence of postpartum metabolic syndrome. Obstet. Gynecol. 2009, 114, 1076–1084. [Google Scholar] [CrossRef]
- Al-Nasiry, S.; Ghossein-Doha, C.; Polman, S.E.; Lemmens, S.; Scholten, R.R.; Heidema, W.M.; Spaan, J.J.; Spaanderman, M.E. Metabolic syndrome after pregnancies complicated by pre-eclampsia or small-for-gestational-age: A retrospective cohort. BJOG 2015, 122, 1818–1823. [Google Scholar] [CrossRef]
- Ahmed, R.; Dunford, J.; Mehran, R.; Robson, S.; Kunadian, V. Pre-eclampsia and future cardiovascular risk among women: A review. J. Am. Coll. Cardiol. 2014, 63, 1815–1822. [Google Scholar] [CrossRef]
- Powe, C.E.; Levine, R.J.; Karumanchi, S.A. Preeclampsia, a disease of the maternal endothelium: The role of antiangiogenic factors and implications for later cardiovascular disease. Circulation 2011, 123, 2856–2869. [Google Scholar] [CrossRef]
- Chen, C.W.; Jaffe, I.Z.; Karumanchi, S.A. Pre-eclampsia and cardiovascular disease. Cardiovasc. Res. 2014, 101, 579–586. [Google Scholar] [CrossRef]
- Barker, D.J.; Osmond, C.; Golding, J.; Kuh, D.; Wadsworth, M.E. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ 1989, 298, 564–567. [Google Scholar] [CrossRef]
- Vikse, B.E.; Irgens, L.M.; Leivestad, T.; Skjaerven, R.; Iversen, B.M. Preeclampsia and the risk of end-stage renal disease. N. Engl. J. Med. 2008, 359, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Ritz, E.; Amann, K.; Koleganova, N.; Benz, K. Prenatal programming-effects on blood pressure and renal function. Nat. Rev. Nephrol. 2011, 7, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.; Park, J.; Gilman-Sachs, A.; Kwak-Kim, J. Immunologic characteristics of preeclampsia, a comprehensive review. Am. J. Reprod. Immunol. 2011, 65, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Chaiworapongsa, T.; Chaemsaithong, P.; Korzeniewski, S.J.; Yeo, L.; Romero, R. Pre-eclampsia part 2: Prediction, prevention and management. Nat. Rev. Nephrol. 2014, 10, 531–540. [Google Scholar] [CrossRef]
- Kelly, R.S.; Croteau-Chonka, D.C.; Dahlin, A.; Mirzakhani, H.; Wu, A.C.; Wan, E.S.; McGeachie, M.J.; Qiu, W.; Sordillo, J.E.; Al-Garawi, A.; et al. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics 2017, 13, 7. [Google Scholar] [CrossRef]
- Maynard, S.E.; Min, J.Y.; Merchan, J.; Lim, K.H.; Li, J.; Mondal, S.; Libermann, T.A.; Morgan, J.P.; Sellke, F.W.; Stillman, I.E.; et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J. Clin. Investig. 2003, 111, 649–658. [Google Scholar] [CrossRef]
- Venkatesha, S.; Toporsian, M.; Lam, C.; Hanai, J.; Mammoto, T.; Kim, Y.M.; Bdolah, Y.; Lim, K.H.; Yuan, H.T.; Libermann, T.A.; et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat. Med. 2006, 12, 642–649. [Google Scholar] [CrossRef]
- Kleinrouweler, C.E.; van Uitert, M.; Moerland, P.D.; Ris-Stalpers, C.; van der Post, J.A.; Afink, G.B. Differentially expressed genes in the pre-eclamptic placenta: A systematic review and meta-analysis. PLoS ONE 2013, 8, e68991. [Google Scholar] [CrossRef]
- Louwen, F.; Muschol-Steinmetz, C.; Reinhard, J.; Reitter, A.; Yuan, J. A lesson for cancer research: Placental microarray gene analysis in preeclampsia. Oncotarget 2012, 3, 759–773. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Yong, H.E.; Melton, P.E.; Johnson, M.P.; Freed, K.A.; Kalionis, B.; Murthi, P.; Brennecke, S.P.; Keogh, R.J.; Moses, E.K. Genome-wide transcriptome directed pathway analysis of maternal pre-eclampsia susceptibility genes. PLoS ONE 2015, 10, e0128230. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Li, C.; Liu, C.X. Immune cell infiltration landscape and immune marker molecular typing in preeclampsia. Bioengineered 2021, 12, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Li, W.; Xiao, J.; Yu, N.; Fan, L.; Sha, M.; Ma, S.; Wu, J.; Chen, S. Integrated analysis of multiple microarray studies to identify novel gene signatures in preeclampsia. Placenta 2021, 105, 104–118. [Google Scholar] [CrossRef]
- He, J.; Liu, K.; Hou, X.; Lu, J. Identification and validation of key non-coding RNAs and mRNAs using co-expression network analysis in pre-eclampsia. Medicine 2021, 100, e25294. [Google Scholar] [CrossRef]
- Chu, X.Y.; Quan, Y.; Zhang, H.Y. Human accelerated genome regions with value in medical genetics and drug discovery. Drug Discov. Today 2020, 25, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Bufill, E.; Blesa, R.; Augusti, J. Alzheimer’s disease: An evolutionary approach. J. Anthropol. Sci. 2013, 91, 135–157. [Google Scholar] [CrossRef]
- Doan, R.N.; Bae, B.I.; Cubelos, B.; Chang, C.; Hossain, A.A.; Al-Saad, S.; Mukaddes, N.M.; Oner, O.; Al-Saffar, M.; Balkhy, S.; et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 2016, 167, 341–354.e12. [Google Scholar] [CrossRef]
- Kallio, M.A.; Tuimala, J.T.; Hupponen, T.; Klemela, P.; Gentile, M.; Scheinin, I.; Koski, M.; Kaki, J.; Korpelainen, E.I. Chipster: User-friendly analysis software for microarray and other high-throughput data. BMC Genom. 2011, 12, 507. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Razick, S.; Magklaras, G.; Donaldson, I.M. iRefIndex: A consolidated protein interaction database with provenance. BMC Bioinform. 2008, 9, 405. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabasi, A.L. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Z.; Sun, M.; Liu, Q.; Wei, M.; Gao, H. Identification of cancer/testis antigen 2 gene as a potential hepatocellular carcinoma therapeutic target by hub gene screening with topological analysis. Oncol. Lett. 2019, 18, 4778–4788. [Google Scholar] [CrossRef]
- Murga-Moreno, J.; Coronado-Zamora, M.; Bodelon, A.; Barbadilla, A.; Casillas, S. PopHumanScan: The online catalog of human genome adaptation. Nucleic Acids Res. 2019, 47, D1080–D1089. [Google Scholar] [CrossRef]
- Sharma, A.; Menche, J.; Huang, C.C.; Ort, T.; Zhou, X.; Kitsak, M.; Sahni, N.; Thibault, D.; Voung, L.; Guo, F.; et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum. Mol. Genet. 2015, 24, 3005–3020. [Google Scholar] [CrossRef]
- Pollard, K.S.; Salama, S.R.; King, B.; Kern, A.D.; Dreszer, T.; Katzman, S.; Siepel, A.; Pedersen, J.S.; Bejerano, G.; Baertsch, R.; et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006, 2, e168. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Terada, T.; Muto, Y. Systems Level Analysis and Identification of Pathways and Key Genes Associated with Delirium. Genes 2020, 11, 1225. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, S.; Umemura, T.; Terada, T.; Muto, Y. Network and Evolutionary Analysis of Human Epigenetic Regulators to Unravel Disease Associations. Genes 2020, 11, 1457. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; de Lange, S.C.; Scholtens, L.H.; Watanabe, K.; Ardesch, D.J.; Jansen, P.R.; Savage, J.E.; Li, L.; Preuss, T.M.; Rilling, J.K.; et al. Genetic mapping and evolutionary analysis of human-expanded cognitive networks. Nat. Commun. 2019, 10, 4839. [Google Scholar] [CrossRef]
- Horvath, S. Weighted Network Analysis: Application in Genomics and Systems Biology; Springer: New York, NY, USA, 2011; pp. 91–121. [Google Scholar]
- Liu, J.; Jing, L.; Tu, X. Weighted gene co-expression network analysis identifies specific modules and hub genes related to coronary artery disease. BMC Cardiovasc. Disord. 2016, 16, 54. [Google Scholar] [CrossRef]
- Buhimschi, I.A.; Nayeri, U.A.; Zhao, G.; Shook, L.L.; Pensalfini, A.; Funai, E.F.; Bernstein, I.M.; Glabe, C.G.; Buhimschi, C.S. Protein misfolding, congophilia, oligomerization, and defective amyloid processing in preeclampsia. Sci. Transl. Med. 2014, 6, 245ra292. [Google Scholar] [CrossRef]
- Mao, X.R.; Crowder, C.M. Protein misfolding induces hypoxic preconditioning via a subset of the unfolded protein response machinery. Mol. Cell. Biol. 2010, 30, 5033–5042. [Google Scholar] [CrossRef]
- Paschen, W.; Mengesdorf, T. Endoplasmic reticulum stress response and neurodegeneration. Cell Calcium 2005, 38, 409–415. [Google Scholar] [CrossRef]
- Jian, B.; Hsieh, C.H.; Chen, J.; Choudhry, M.; Bland, K.; Chaudry, I.; Raju, R. Activation of endoplasmic reticulum stress response following trauma-hemorrhage. Biochim. Biophys. Acta 2008, 1782, 621–626. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, H.; Yang, Y.; Guo, F.; Yu, B.; Su, Z. Trophoblast Cell Subtypes and Dysfunction in the Placenta of Individuals with Preeclampsia Revealed by SingleCell RNA Sequencing. Mol. Cells 2022, 45, 317–328. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Tsarev, A.A.; Vashukova, E.S.; Maksiutenko, E.M.; Kovalenko, L.V.; Belotserkovtseva, L.D.; Glotov, A.S. A Data-Driven Review of the Genetic Factors of Pregnancy Complications. Int. J. Mol. Sci. 2020, 21, 3384. [Google Scholar] [CrossRef] [PubMed]
- Benny, P.A.; Alakwaa, F.M.; Schlueter, R.J.; Lassiter, C.B.; Garmire, L.X. A review of omics approaches to study preeclampsia. Placenta 2020, 92, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.L.; Swanson, W.J. Positive selection in the human genome: From genome scans to biological significance. Annu. Rev. Genom. Hum. Genet. 2008, 9, 143–160. [Google Scholar] [CrossRef]
- Romagnoli, A.; D’Agostino, M.; Ardiccioni, C.; Maracci, C.; Motta, S.; La Teana, A.; Di Marino, D. Control of the eIF4E activity: Structural insights and pharmacological implications. Cell Mol. Life Sci. 2021, 78, 6869–6885. [Google Scholar] [CrossRef]
- Carroll, M.; Borden, K.L. The oncogene eIF4E: Using biochemical insights to target cancer. J. Interferon Cytokine Res. 2013, 33, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef]
- Kao, S.H.; Cheng, W.C.; Wang, Y.T.; Wu, H.T.; Yeh, H.Y.; Chen, Y.J.; Tsai, M.H.; Wu, K.J. Regulation of miRNA Biogenesis and Histone Modification by K63-Polyubiquitinated DDX17 Controls Cancer Stem-like Features. Cancer Res. 2019, 79, 2549–2563. [Google Scholar] [CrossRef]
- Dardenne, E.; Espinoza, M.P.; Fattet, L.; Germann, S.; Lambert, M.P.; Neil, H.; Zonta, E.; Mortada, H.; Gratadou, L.; Deygas, M.; et al. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014, 7, 1900–1913. [Google Scholar] [CrossRef]
- Lee, J.H.; Jeong, S.A.; Khadka, P.; Hong, J.; Chung, I.K. Involvement of SRSF11 in cell cycle-specific recruitment of telomerase to telomeres at nuclear speckles. Nucleic Acids Res. 2015, 43, 8435–8451. [Google Scholar] [CrossRef]
- Wang, B.; Ma, A.; Zhang, L.; Jin, W.L.; Qian, Y.; Xu, G.; Qiu, B.; Yang, Z.; Liu, Y.; Xia, Q.; et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 2015, 6, 8704. [Google Scholar] [CrossRef]
- Luo, G.; Hu, N.; Xia, X.; Zhou, J.; Ye, C. RPN11 deubiquitinase promotes proliferation and migration of breast cancer cells. Mol. Med. Rep. 2017, 16, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Z.; Bi, F.; Yang, Q. Deubiquitinase PSMD14 promotes ovarian cancer progression by decreasing enzymatic activity of PKM2. Mol. Oncol. 2021, 15, 3639–3658. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, C.; Liu, Y.; Wang, N.; Gao, S.; Qiu, S.; Wang, Z.; Ding, J.; Zhang, L.; Wang, H.; et al. Identifying key genes and drug screening for preeclampsia based on gene expression profiles. Oncol. Lett. 2020, 20, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Prohaska, A.; Racimo, F.; Schork, A.J.; Sikora, M.; Stern, A.J.; Ilardo, M.; Allentoft, M.E.; Folkersen, L.; Buil, A.; Moreno-Mayar, J.V.; et al. Human Disease Variation in the Light of Population Genomics. Cell 2019, 177, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Byars, S.G.; Voskarides, K. Antagonistic Pleiotropy in Human Disease. J. Mol. Evol. 2020, 88, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Corbett, S.; Courtiol, A.; Lummaa, V.; Moorad, J.; Stearns, S. The transition to modernity and chronic disease: Mismatch and natural selection. Nat. Rev. Genet. 2018, 19, 419–430. [Google Scholar] [CrossRef]
- Robillard, P.Y.; Dekker, G.; Iacobelli, S.; Chaouat, G. An essay of reflection: Why does preeclampsia exist in humans, and why are there such huge geographical differences in epidemiology? J. Reprod. Immunol. 2016, 114, 44–47. [Google Scholar] [CrossRef]
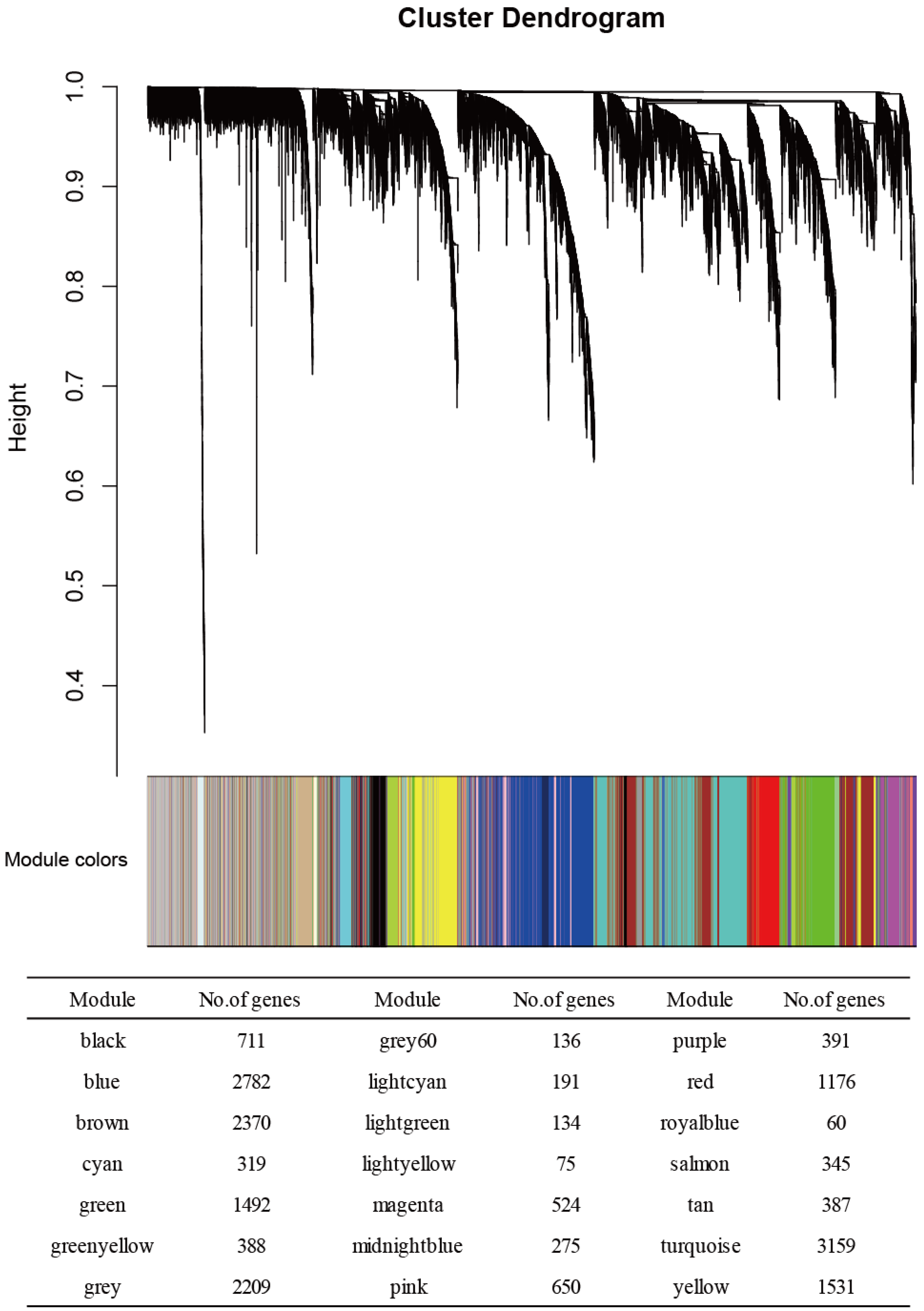
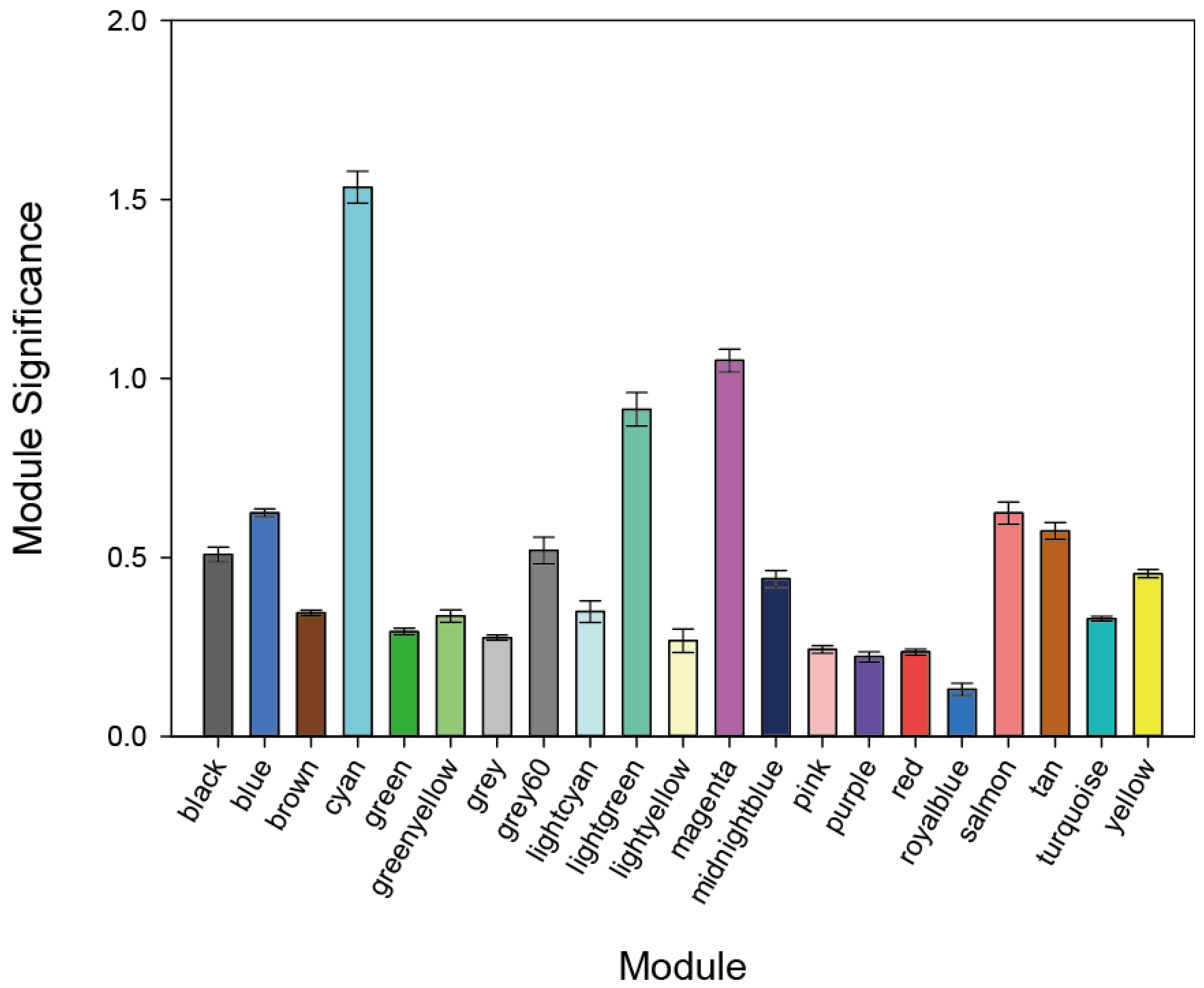
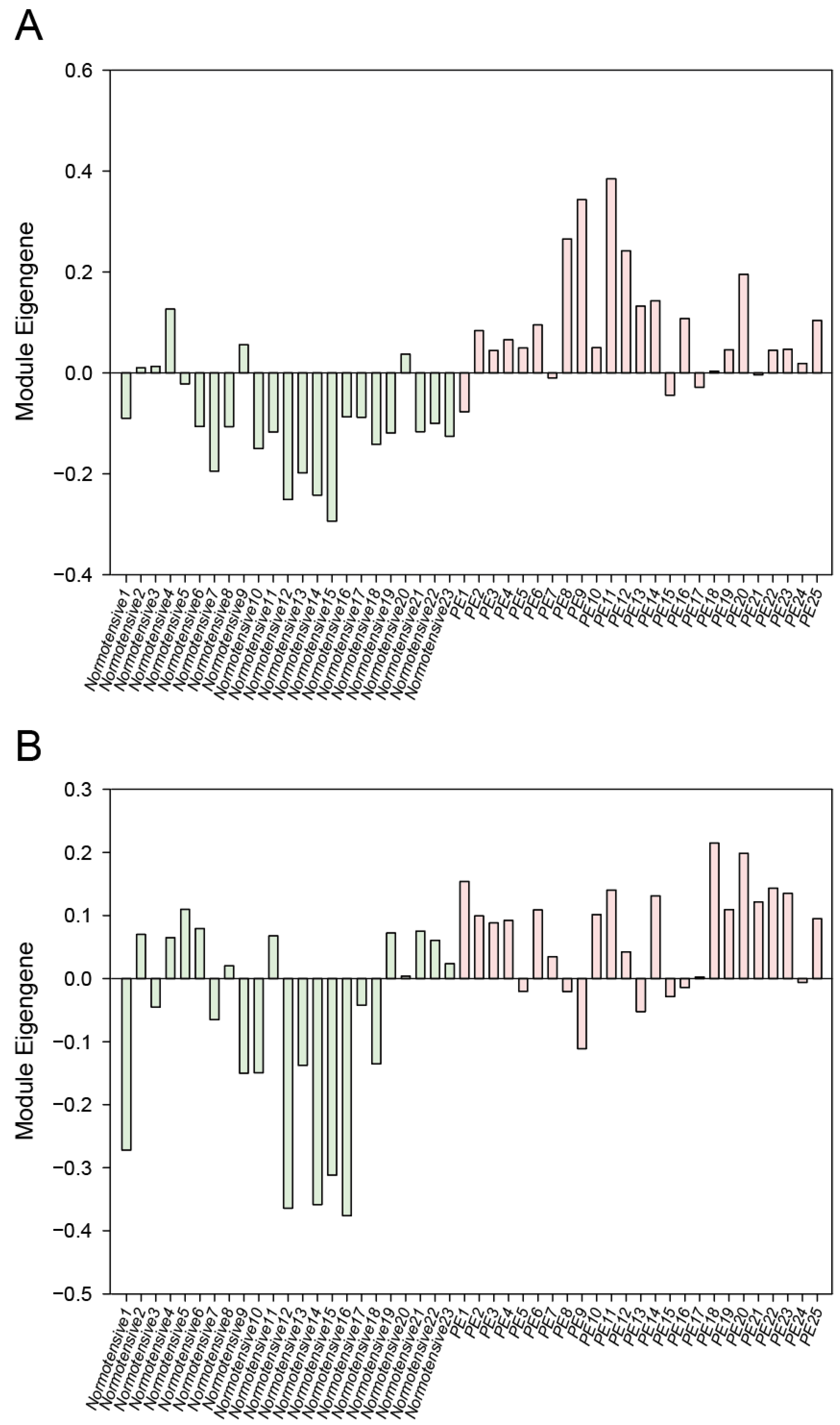
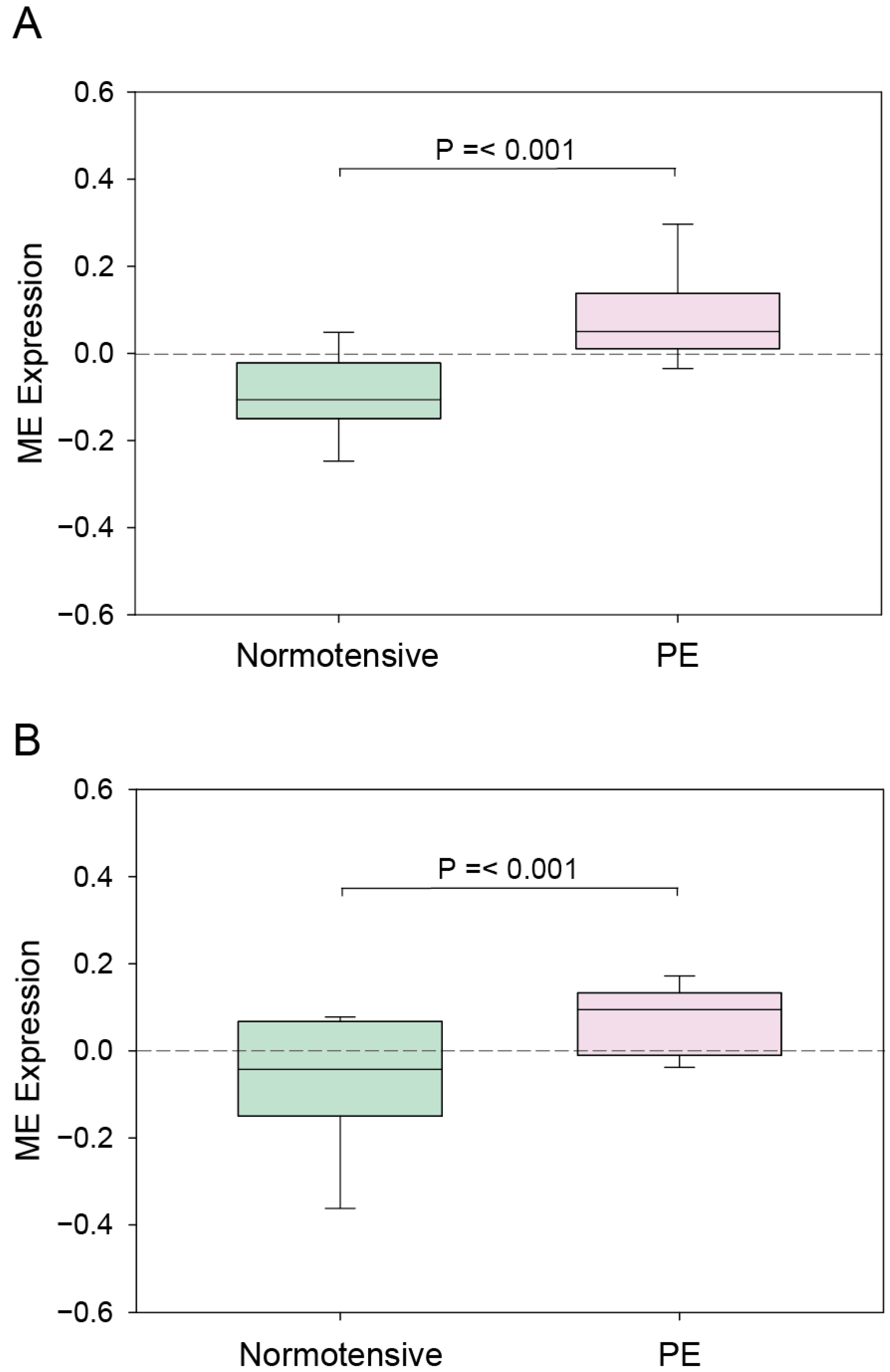
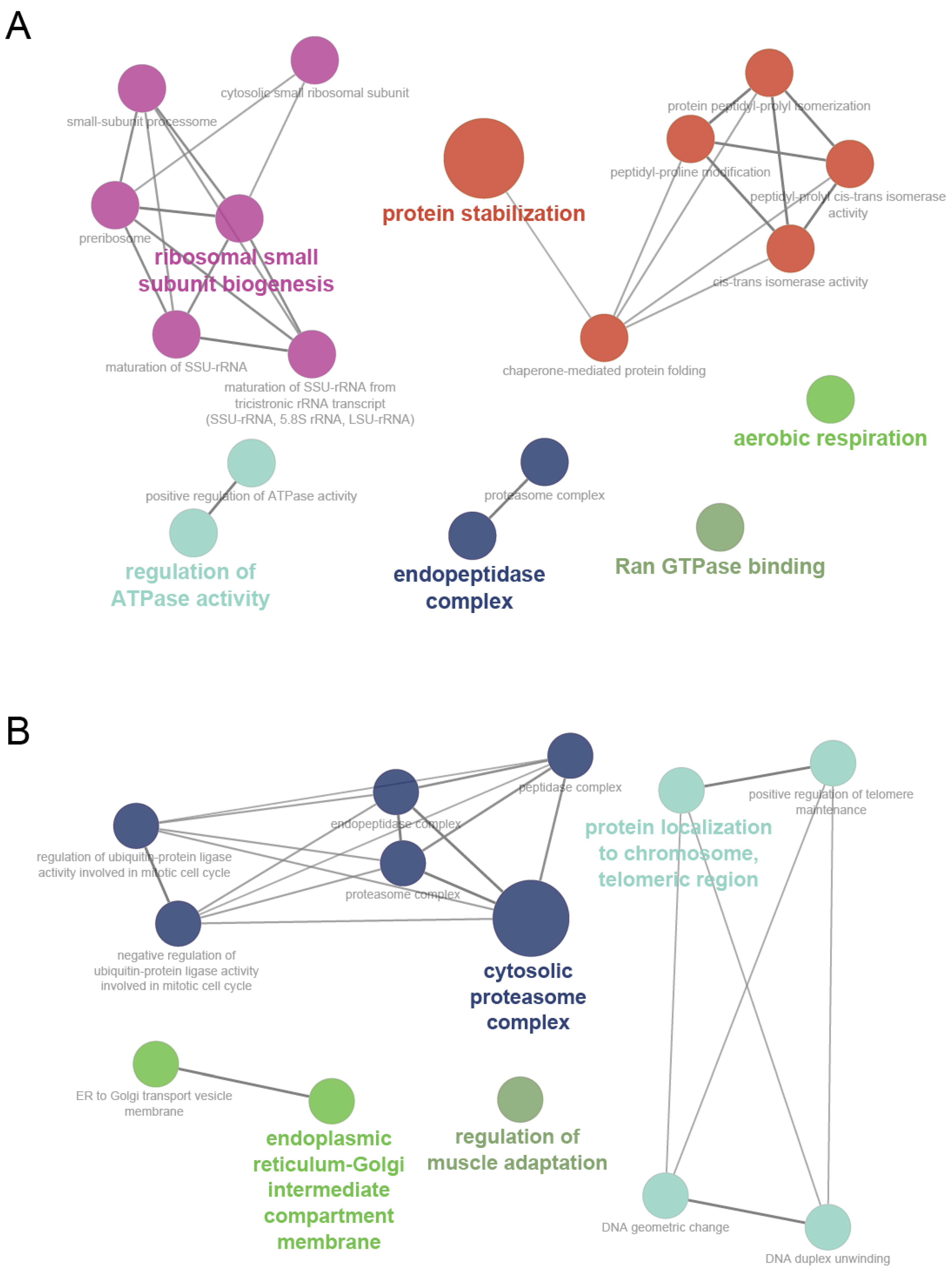
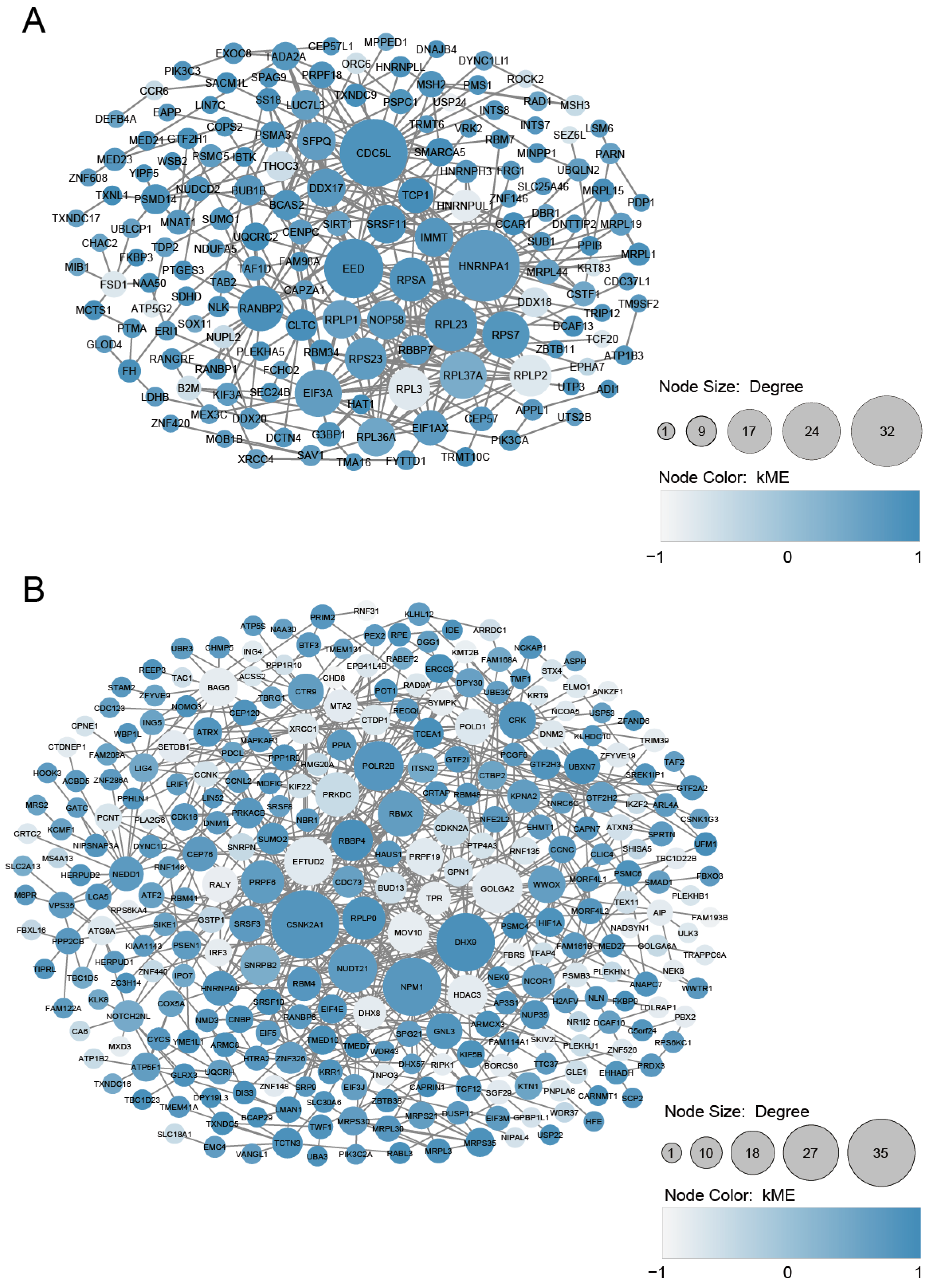
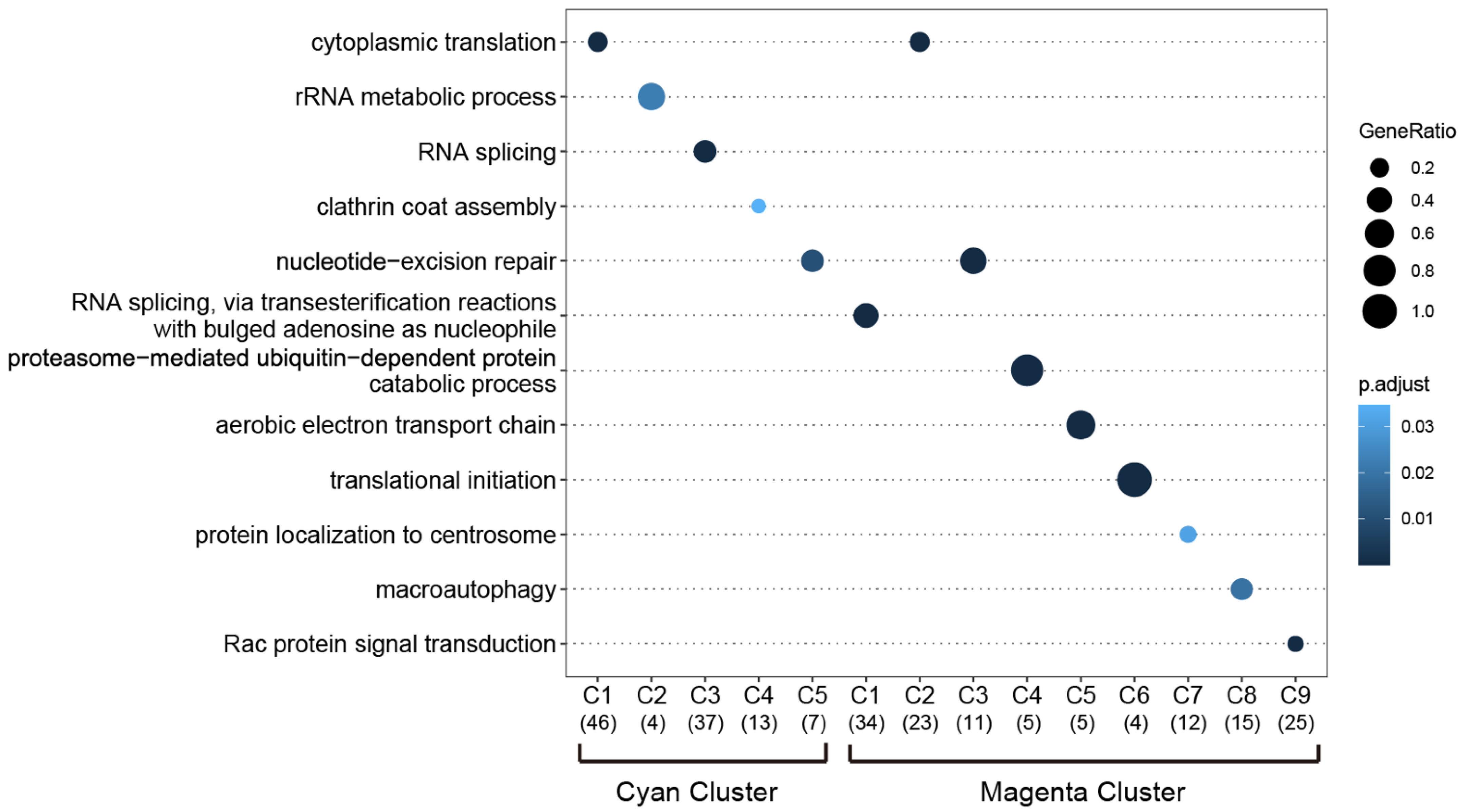
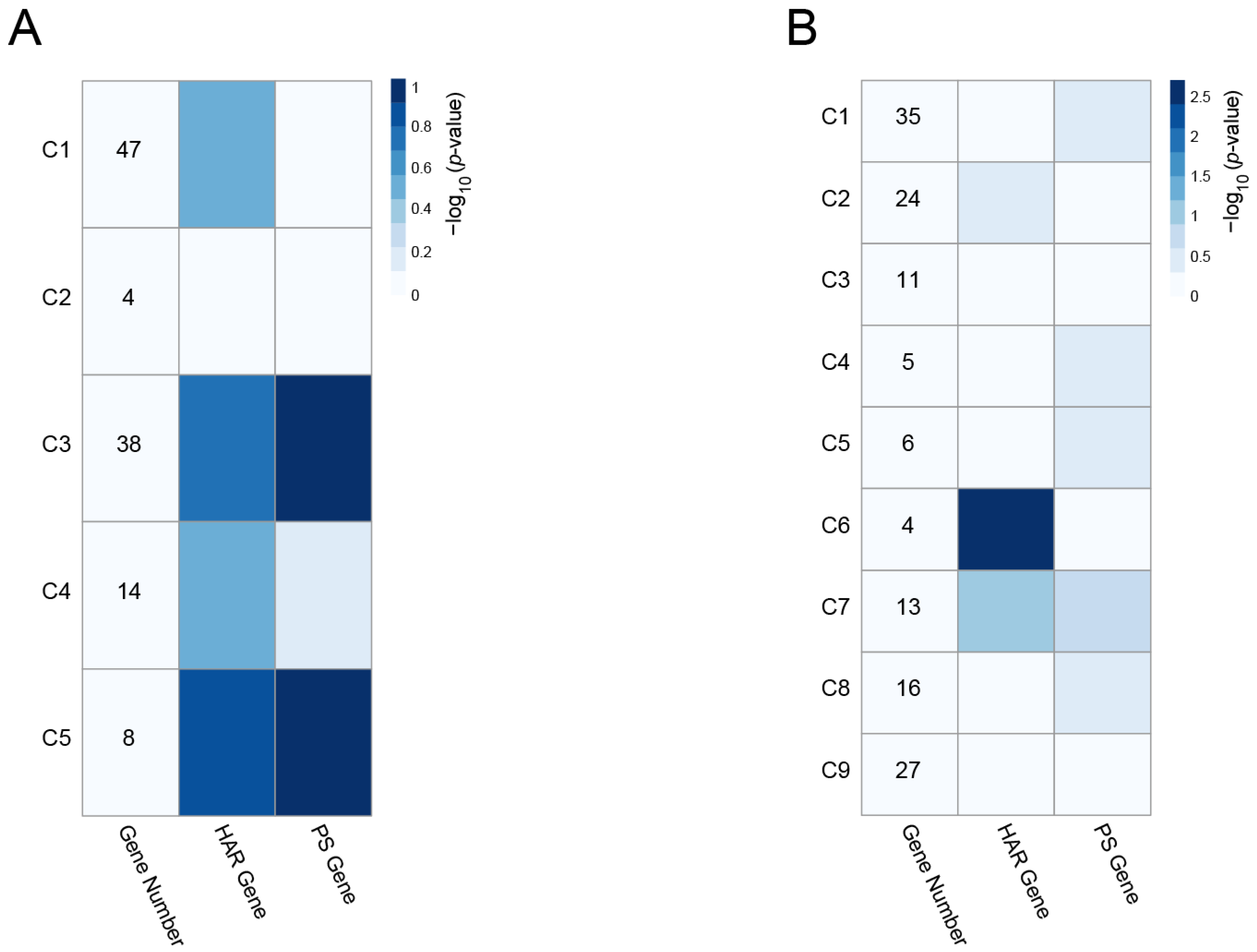
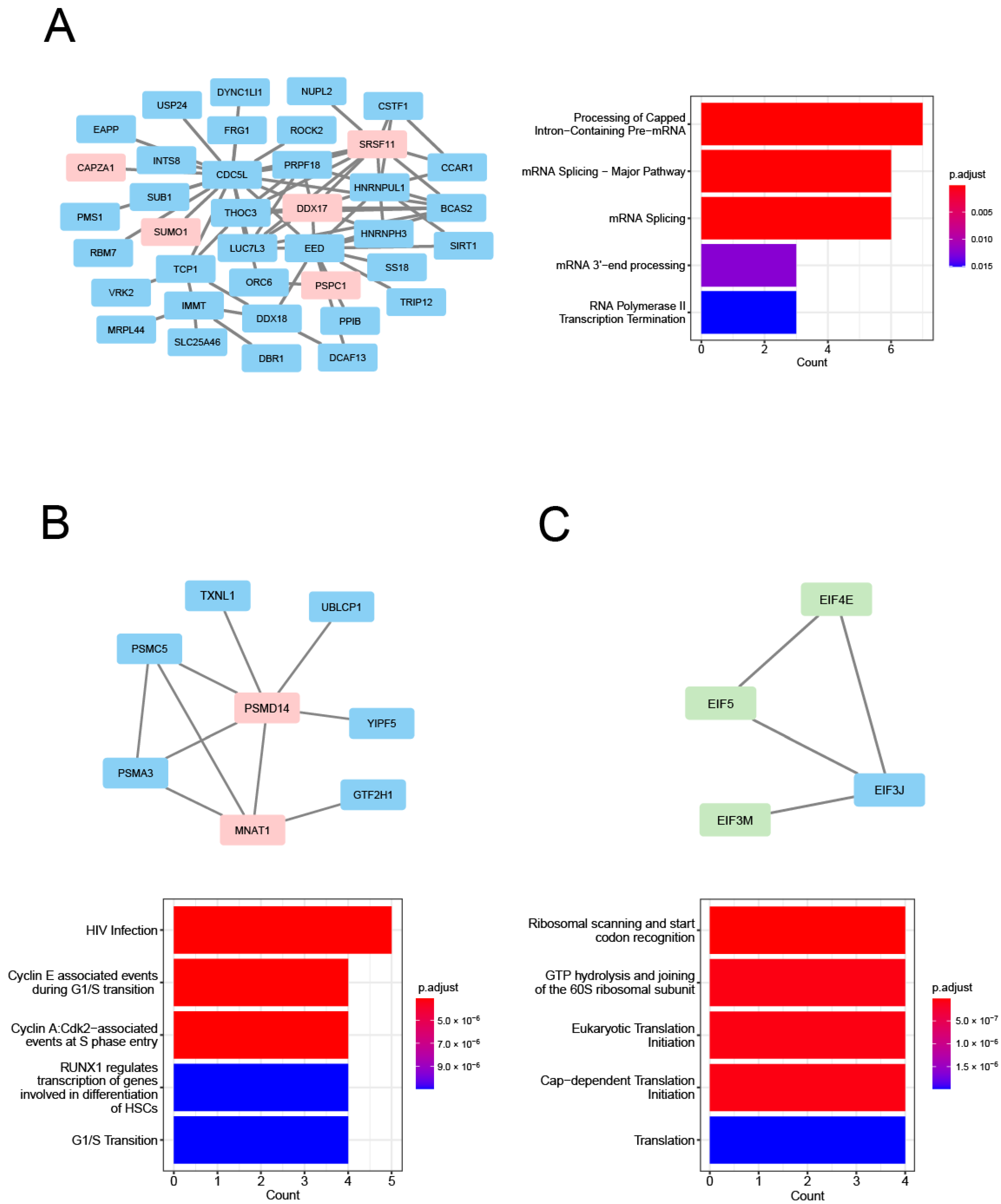
| Module Term | p−Value | Count | Genes |
|---|---|---|---|
| Cyan | |||
| GO:0050821-protein stabilization | 2.66 × 10−5 | 7 | [ATP1B3, NLK, PPIB, PTGES3, RPS7, SUMO1, TCP1] |
| GO:0061077-chaperone-mediated protein folding | 3.93 × 10−4 | 4 | [FKBP3, PPIB, PTGES3, TCP1] |
| GO:0042274-ribosomal small subunit biogenesis | 4.39 × 10−4 | 4 | [DCAF13, RPS7, RPSA, UTP3] |
| GO:0008536-Ran GTPase binding | 6.45 × 10−4 | 3 | [RANBP1, RANBP2, RANGRF] |
| GO:0030684-preribosome | 6.58 × 10−4 | 4 | [DCAF13, RPS7, RPSA, UTP3] |
| GO:0000462-maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 7.64 × 10−4 | 3 | [DCAF13, RPSA, UTP3] |
| GO:0032040-small-subunit processome | 8.28 × 10−4 | 3 | [DCAF13, RPS7, UTP3] |
| GO:0003755-peptidyl-prolyl cis-trans isomerase activity | 1.20 × 10−3 | 3 | [FKBP3, PPIB, RANBP2] |
| GO:0000413-protein peptidyl-prolyl isomerization | 1.20 × 10−3 | 3 | [FKBP3, PPIB, RANBP2] |
| GO:0016859-cis-trans isomerase activity | 1.38 × 10−3 | 3 | [FKBP3, PPIB, RANBP2] |
| GO:0022627-cytosolic small ribosomal subunit | 2.11 × 10−3 | 3 | [MCTS1, RPS7, RPSA] |
| GO:0030490-maturation of SSU-rRNA | 2.23 × 10−3 | 3 | [DCAF13, RPSA, UTP3] |
| GO:0032781-positive regulation of ATPase activity | 2.49 × 10−3 | 3 | [ATP1B3, DNAJB4, SUMO1] |
| GO:0018208-peptidyl-proline modification | 2.90 × 10−3 | 3 | [FKBP3, PPIB, RANBP2] |
| GO:0000502-proteasome complex | 4.98 × 10−3 | 3 | [PSMA3, PSMD14, TXNL1] |
| GO:1905369-endopeptidase complex | 5.18 × 10−3 | 3 | [PSMA3, PSMD14, TXNL1] |
| GO:0009060-aerobic respiration | 5.61 × 10−3 | 3 | [FH, SDHD, UQCRC2] |
| GO:0043462-regulation of ATPase activity | 6.28 × 10−3 | 3 | [ATP1B3, DNAJB4, SUMO1] |
| Magenta | |||
| GO:0031597-cytosolic proteasome complex | 3.33 × 10−5 | 3 | [IDE, PSMC4, PSMC6] |
| GO:0070198-protein localization to chromosome, telomeric region | 4.90 × 10−4 | 3 | [ATRX, GNL3, POT1] |
| GO:0032508-DNA duplex unwinding | 6.27 × 10−4 | 4 | [ATRX, DHX9, GTF2H3, POT1] |
| GO:0032392-DNA geometric change | 1.03 × 10−3 | 4 | [ATRX, DHX9, GTF2H3, POT1] |
| GO:1905368-peptidase complex | 1.26 × 10−3 | 4 | [IDE, PSMC4, PSMC6, USP22] |
| GO:0032206-positive regulation of telomere maintenance | 2.49 × 10−3 | 3 | [ATRX, GNL3, POT1] |
| GO:0012507-ER to Golgi transport vesicle membrane | 4.03 × 10−3 | 3 | [LMAN1, TMED10, TMED7] |
| GO:0000502-proteasome complex | 4.98 × 10−3 | 3 | [IDE, PSMC4, PSMC6] |
| GO:1905369-endopeptidase complex | 5.18 × 10−3 | 3 | [IDE, PSMC4, PSMC6] |
| GO:0033116-endoplasmic reticulum-Golgi intermediate compartment membrane | 5.39 × 10−3 | 3 | [LMAN1, TMED10, TMED7] |
| GO:0051436-negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.83 × 10−3 | 3 | [ANAPC7, PSMC4, PSMC6] |
| GO:0051439-regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 6.05 × 10−3 | 3 | [ANAPC7, PSMC4, PSMC6] |
| GO:0043502-regulation of muscle adaptation | 6.28 × 10−3 | 3 | [GLRX3, GTF2I, TWF1] |
| Scheme | Gene Full Name | Module | Cluster | Degree | Betweenness | HAR Gene | PS Gene |
|---|---|---|---|---|---|---|---|
| DDX17 | DEAD-Box Helicase 17 | cyan | C3 | 13 | 533.6621 | O | |
| SRSF11 | Serine and Arginine Rich Splicing Factor 11 | cyan | C3 | 13 | 785.37203 | O | |
| PSPC1 | Paraspeckle Component 1 | cyan | C3 | 5 | 364.12826 | O | |
| SUMO1 | Small Ubiquitin-like Modifier 1 | cyan | C3 | 4 | 620 | O | |
| CAPZA1 | Capping Actin Protein of Muscle Z-Line | cyan | C3 | 2 | 206.9429 | O | |
| Subunit α 1 | |||||||
| PSMD14 | Proteasome 26S Subunit, Non-ATPase 14 | cyan | C5 | 8 | 2130.39568 | O | O |
| MNAT1 | Menage A Trois 1 | cyan | C5 | 5 | 226.87372 | O | |
| EIF4E | Eukaryotic Translation Initiation Factor 4E | magenta | C6 | 7 | 354.64491 | O | |
| EIF5 | Eukaryotic Translation Initiation Factor 5 | magenta | C6 | 4 | 48.22838 | O | |
| EIF3M | Eukaryotic Translation Initiation Factor 3 Subunit M | magenta | C6 | 4 | 617.89841 | O |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondoh, K.; Akahori, H.; Muto, Y.; Terada, T. Identification of Key Genes and Pathways Associated with Preeclampsia by a WGCNA and an Evolutionary Approach. Genes 2022, 13, 2134. https://doi.org/10.3390/genes13112134
Kondoh K, Akahori H, Muto Y, Terada T. Identification of Key Genes and Pathways Associated with Preeclampsia by a WGCNA and an Evolutionary Approach. Genes. 2022; 13(11):2134. https://doi.org/10.3390/genes13112134
Chicago/Turabian StyleKondoh, Kuniyo, Hiromichi Akahori, Yoshinori Muto, and Tomoyoshi Terada. 2022. "Identification of Key Genes and Pathways Associated with Preeclampsia by a WGCNA and an Evolutionary Approach" Genes 13, no. 11: 2134. https://doi.org/10.3390/genes13112134
APA StyleKondoh, K., Akahori, H., Muto, Y., & Terada, T. (2022). Identification of Key Genes and Pathways Associated with Preeclampsia by a WGCNA and an Evolutionary Approach. Genes, 13(11), 2134. https://doi.org/10.3390/genes13112134






