Novel Insights into MEG3/miR664a-3p/ADH4 Axis and Its Possible Role in Hepatocellular Carcinoma from an in Silico Perspective
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of lncRNA, miR-664a, and ADH4 Sequences
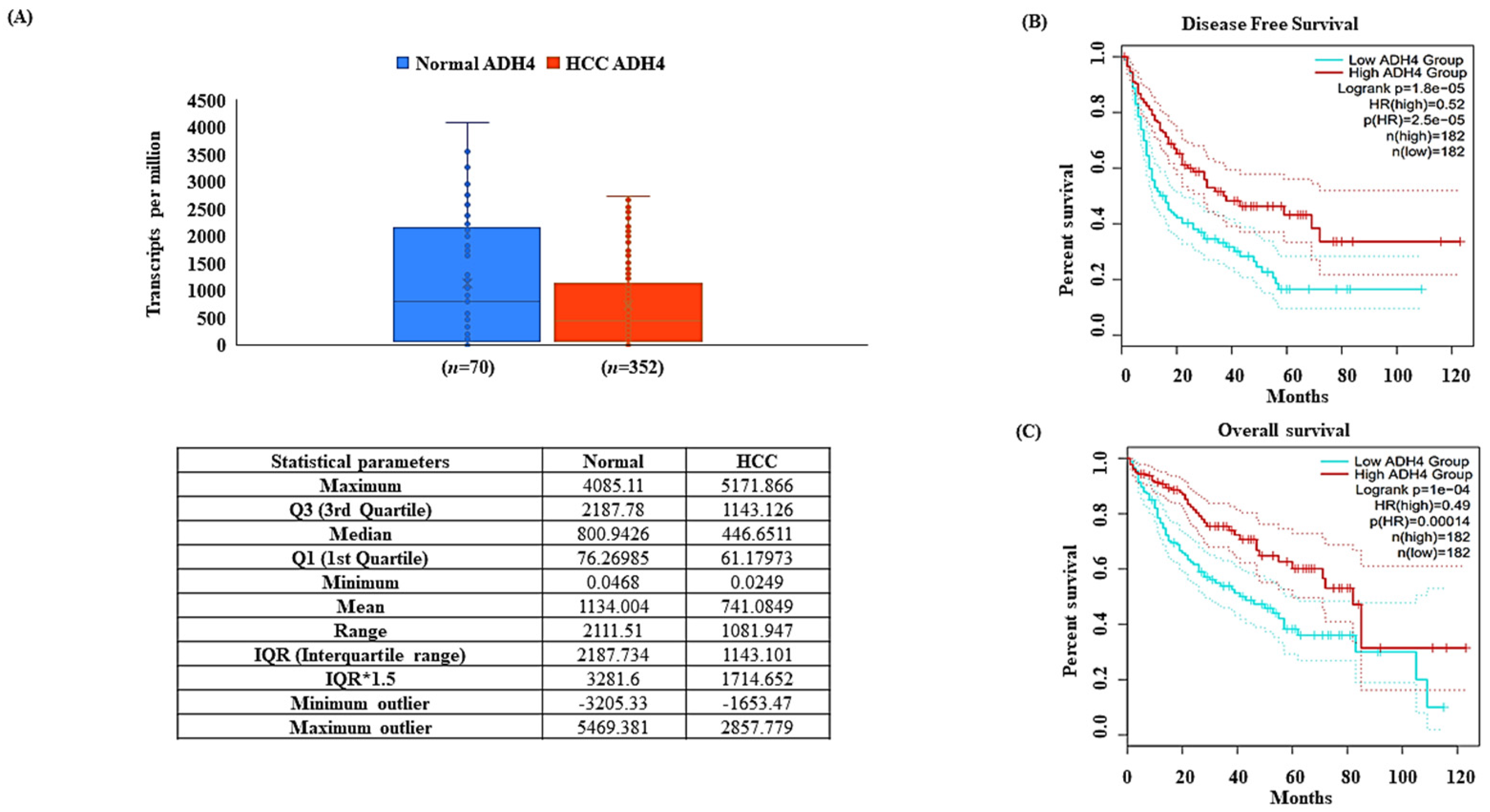
2.2. Expression of ADH4 in HCC
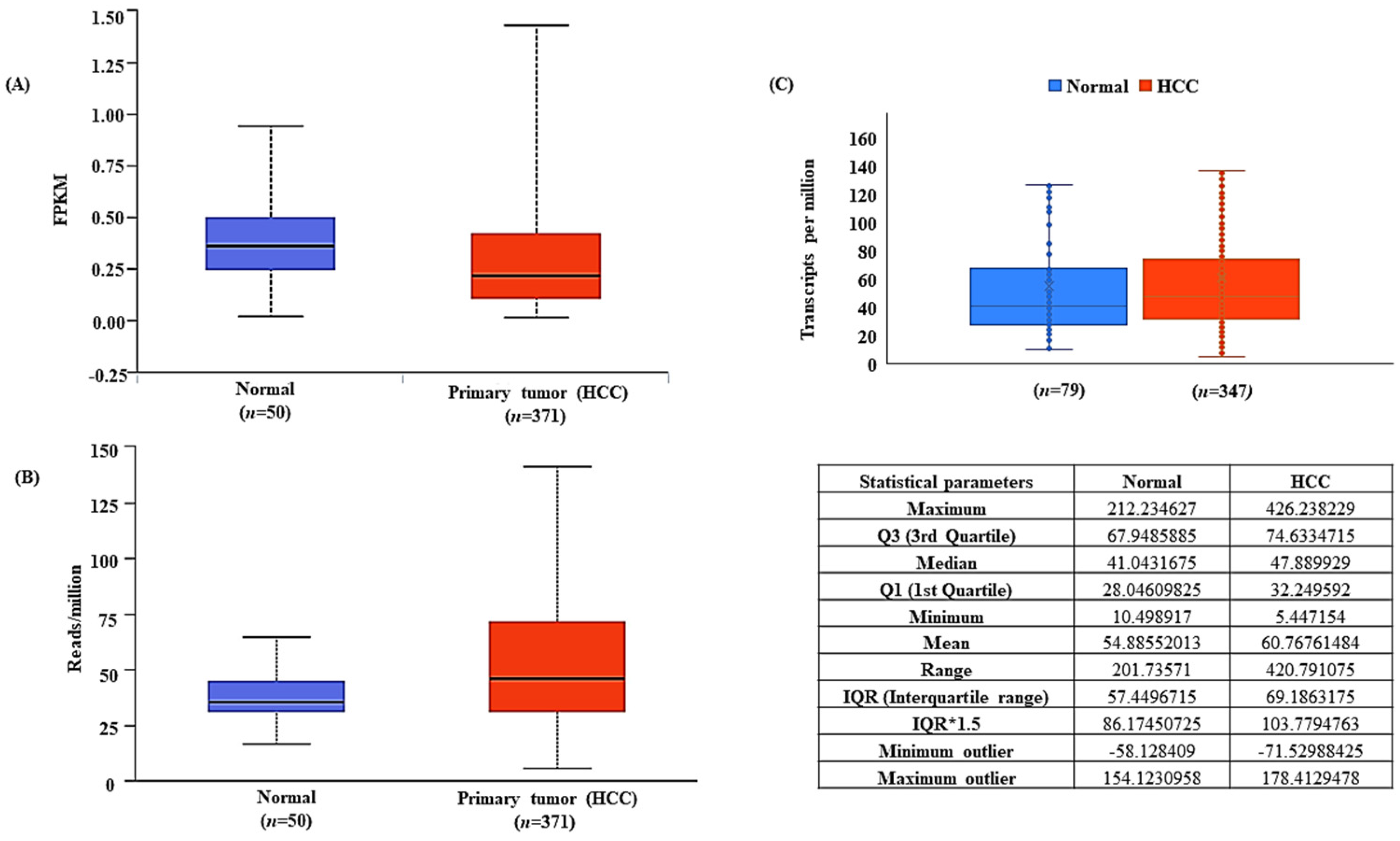
2.3. Differential Expression of miR-664a-3p and MEG3 in HCC
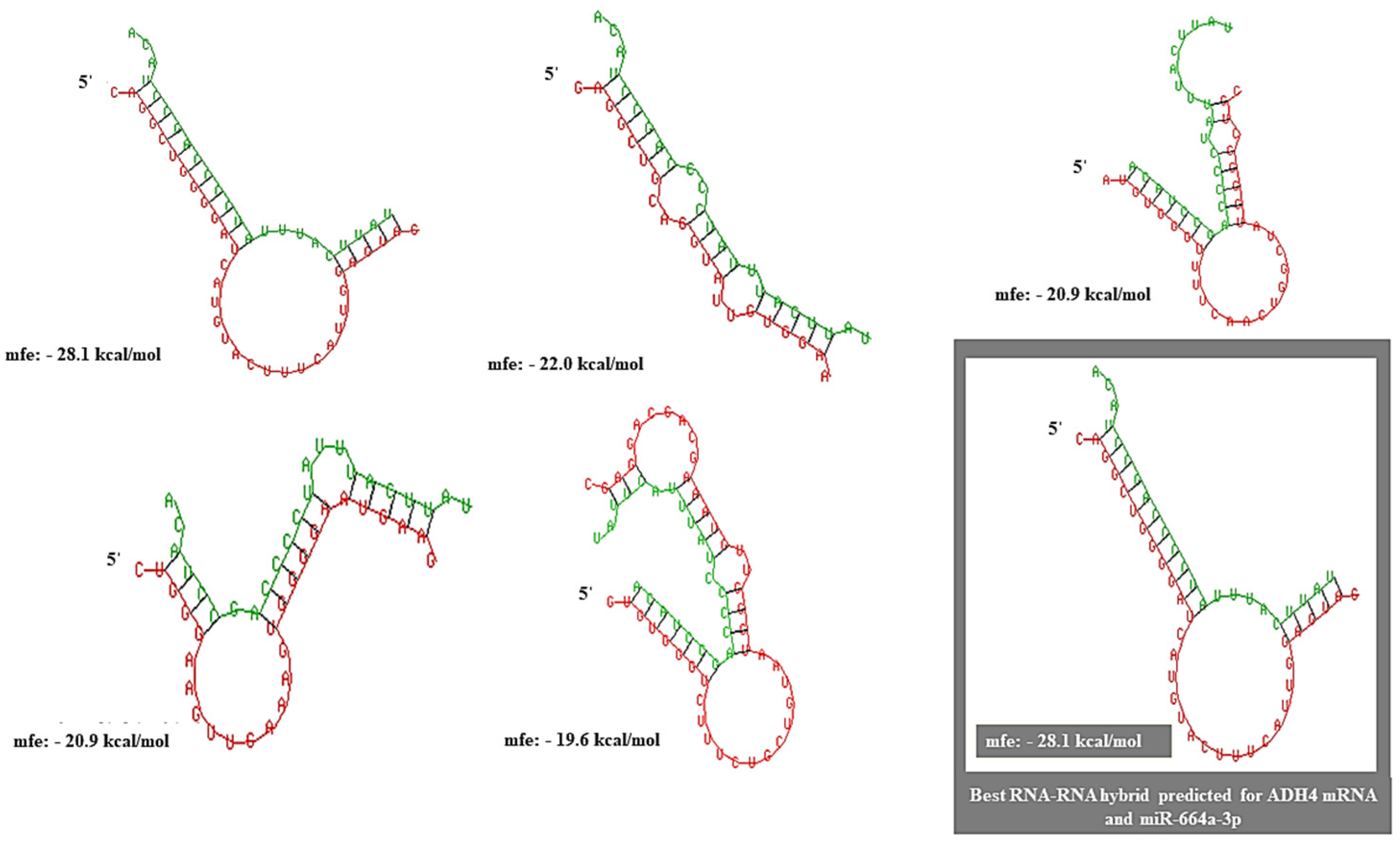
2.4. Construction of miRNA–mRNA RNA Heteroduplex Hybrids
2.5. Construction of lncRNA–miRNA RNA Heteroduplex Hybrids
2.6. Predicting the Accessibility of Binding Regions of MEG3-miRNA RNA Heteroduplex
3. Results
3.1. ADH4 Is Downregulated in HCC
3.2. LncRNA-MEG3 Is Downregulated and miR-664a-3p Is Upregulated in HCC
3.3. ADH4 Is Targeted by miR-664a-3p in HCC
3.4. MEG3 Acts as a Competitive Endogenous Sponge for miR-664a-3p
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konyn, P.; Ahmed, A.; Kim, D. Current epidemiology in hepatocellular carcinoma. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tian, Z. HBV-Induced Immune Imbalance in the Development of HCC. Front. Immunol. 2019, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Gao, Y.; He, Y.; Hooper, J.D.; Yang, P. HBV induced hepatocellular carcinoma and related potential immunotherapy. Pharmacol. Res. 2020, 159, 104992. [Google Scholar] [CrossRef]
- Jiang, Y.; Han, Q.; Zhao, H.; Zhang, J. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.A.; Ijaz, B.; Daud, M.; Tariq, S.; Nadeem, T.; Husnain, T. Interplay of Wnt β-catenin pathway and miRNAs in HBV pathogenesis leading to HCC. Clin. Res. Hepatol. Gastroenterol. 2019, 43, 373–386. [Google Scholar] [CrossRef] [PubMed]
- De Battista, D.; Zamboni, F.; Gerstein, H.; Sato, S.; Markowitz, T.E.; Lack, J.; Engle, R.E.; Farci, P. Molecular Signature and Immune Landscape of HCV-Associated Hepatocellular Carcinoma (HCC): Differences and Similarities with HBV-HCC. J. Hepatocell. Carcinoma 2021, 8, 1399–1413. [Google Scholar] [CrossRef]
- Hu, J.; Liu, K.; Luo, J. HIV-HBV and HIV-HCV Coinfection and Liver Cancer Development. Cancer Treat. Res. 2019, 177, 231–250. [Google Scholar] [CrossRef]
- Foerster, F.; Gairing, S.J.; Müller, L.; Galle, P.R. NAFLD-driven HCC: Safety and efficacy of current and emerging treatment options. J. Hepatol. 2022, 76, 446–457. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef]
- Ioannou, G.N. Epidemiology and risk-stratification of NAFLD-associated HCC. J. Hepatol. 2021, 75, 1476–1484. [Google Scholar] [CrossRef]
- Michelotti, G.A.; Machado, M.V.; Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, D.S.; An, T.H.; Park, H.-J.; Kim, W.K.; Bae, K.-H.; Oh, K.-J. Metabolic Spectrum of Liver Failure in Type 2 Diabetes and Obesity: From NAFLD to NASH to HCC. Int. J. Mol. Sci. 2021, 22, 4495. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-S.; Ban, H.S.; Hur, K.; Cho, H.-S. The Epigenetic Regulation of HCC Metastasis. Int. J. Mol. Sci. 2018, 19, 3978. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Taheri, M. Maternally expressed gene 3 (MEG3): A tumor suppressor long non coding RNA. Biomed. Pharmacother. 2019, 118, 109129. [Google Scholar] [CrossRef]
- Al-Rugeebah, A.; Alanazi, M.; Parine, N.R. MEG3: An Oncogenic Long Non-coding RNA in Different Cancers. Pathol. Oncol. Res. 2019, 25, 859–874. [Google Scholar] [CrossRef]
- Mahpour, A.; Mullen, A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. Innov. Hepatol. 2021, 3, 100177. [Google Scholar] [CrossRef]
- He, Y.; Luo, Y.; Liang, B.; Ye, L.; Lu, G.; He, W. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget 2017, 8, 73282–73295. [Google Scholar] [CrossRef]
- Xu, X.; Zhong, Z.; Shao, Y.; Yi, Y. Prognostic Value of MEG3 and Its Correlation With Immune Infiltrates in Gliomas. Front. Genet. 2021, 12, 679097. [Google Scholar] [CrossRef]
- Astuti, D.; Latif, F.; Wagner, K.; Gentle, D.; Cooper, W.N.; Catchpoole, D.; Grundy, R.; Ferguson-Smith, A.C.; Maher, E.R. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: Analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br. J. Cancer 2005, 92, 1574–1580. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Mehta, K.R.; Danila, D.C.; Scolavino, S.; Johnson, S.R.; Klibanski, A. A Pituitary-Derived MEG3 Isoform Functions as a Growth Suppressor in Tumor Cells. J. Clin. Endocrinol. Metab. 2003, 88, 5119–5126. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 Non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Wu, L.; Guo, Y.; Li, G.; Xiang, M.; Liu, L.; Zhan, H.; Zhou, X.; Tan, H. LncRNA MEG3 contributes to adenosine-induced cytotoxicity in hepatoma HepG2 cells by downregulated ILF3 and autophagy inhibition via regulation PI3K-AKT-mTOR and beclin-1 signaling pathway. J. Cell. Biochem. 2019, 120, 18172–18185. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-X.; Deng, W.; Zhou, X.-T.; Chen, R.-P.; Xiang, M.-Q.; Guo, Y.-T.; Pu, Z.-J.; Li, R.; Wang, G.-F.; Wu, L.-F. The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells. Int. J. Oncol. 2016, 48, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Santhekadur, P.K.; Kumar, D.P. RISC assembly and post-transcriptional gene regulation in Hepatocellular Carcinoma. Genes Dis. 2019, 7, 199–204. [Google Scholar] [CrossRef]
- Yoo, B.K.; Santhekadur, P.K.; Gredler, R.; Chen, D.; Emdad, L.; Bhutia, S.; Pannell, L.; Fisher, P.B.; Sarkar, D. Increased RNA-Induced Silencing Complex (RISC) Activity Contributes to Hepatocellular Carcinoma. Hepatology 2011, 53, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Cho, M.E.; Li, T.W.H.; Peng, H.; Ko, K.S.; Mato, J.M.; Lu, S.C. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J. Clin. Investig. 2013, 123, 285–298. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Z.; Zhang, T.; Wang, M.; Xu, R.; Qin, S.; Zhang, S. Overexpression of miR-664 is associated with poor overall survival and accelerates cell proliferation, migration and invasion in hepatocellular carcinoma. OncoTargets Ther. 2019, 12, 2373–2381. [Google Scholar] [CrossRef]
- Edenberg, H.J. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2007, 30, 5–13. [Google Scholar]
- Li, H.; Toth, E.; Cherrington, N.J. Alcohol Metabolism in the Progression of Human Nonalcoholic Steatohepatitis. Toxicol. Sci. 2018, 164, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.-R.; Zhang, M.-Y.; Rao, H.-L.; Pu, H.-Y.; Zhang, H.-Z.; Wang, H.-Y. Identification of ADH4 as a novel and potential prognostic marker in hepatocellular carcinoma. Med. Oncol. 2012, 29, 2737–2743. [Google Scholar] [CrossRef]
- Hong, L.; Zhou, Y.; Xie, X.; Wu, W.; Shi, C.; Lin, H.; Shi, Z. A stemness-based eleven-gene signature correlates with the clinical outcome of hepatocellular carcinoma. BMC Cancer 2021, 21, 716. [Google Scholar] [CrossRef] [PubMed]
- Jelski, W.; Zalewski, B.; Szmitkowski, M. Alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase (ALDH) activity in the sera of patients with liver cancer. J. Clin. Lab. Anal. 2008, 22, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. Publ. Protein Soc. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- GEPIA: A web Server for Cancer and Normal Gene Expression Profiling and interactive Analyses. Available online: https://academic.oup.com/nar/article/45/W1/W98/3605636 (accessed on 22 June 2021).
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Fast and Effective Prediction of microRNA/Target Duplexes. Available online: https://rnajournal.cshlp.org/content/10/10/1507.abstract (accessed on 17 October 2022).
- Freiburg RNA Tools: A Central Online Resource for RNA-Focused Research and Teaching. Available online: https://pubmed.ncbi.nlm.nih.gov/29788132/ (accessed on 17 October 2022).
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Lv, Y.; Zhang, C.; Guo, S. LncRNA meg3 suppresses hepatocellular carcinoma in vitro and vivo studies. Am. J. Transl. Res. 2019, 11, 4089–4099. [Google Scholar]
- Mohammed, S.R.; Shaker, O.G.; Mohamed, M.M.; Abdelhafez Mostafa, M.N.; Gaber, S.N.; Ali, D.Y.; Hussein, H.A.; Elebiary, A.M.; Abonar, A.A.; Eid, H.M.; et al. The emerging role of lncRNA MEG3 and MEG3 rs7158663 in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Gholipour, M.; Hussen, B.M.; Taheri, M. The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 649107. [Google Scholar] [CrossRef]
- Gailhouste, L.; Liew, L.C.; Yasukawa, K.; Hatada, I.; Tanaka, Y.; Kato, T.; Nakagama, H.; Ochiya, T. MEG3-derived miR-493-5p overcomes the oncogenic feature of IGF2-miR-483 loss of imprinting in hepatic cancer cells. Cell Death Dis. 2019, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, S.; Ye, F.; Shen, Y.; Tie, Y.; Zhu, J.; Wei, L.; Jin, Y.; Fu, H.; Wu, Y.; et al. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PLoS ONE 2015, 10, e0139790. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, J.Y.; Zhong, Y.; Xie, L.; Li, J.S. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz. J. Med. Biol. Res. 2019, 52, e8631. [Google Scholar] [CrossRef]
- Li, M.; Liao, H.; Wu, J.; Chen, B.; Pang, R.; Huang, J.; Zhu, Y. Long noncoding RNA matrilineal expression gene 3 inhibits hepatocellular carcinoma progression by targeting microRNA-5195-3p and regulating the expression of forkhead box O1. Bioengineered 2021, 12, 12880–12890. [Google Scholar] [CrossRef]
- Liu, X.; Li, T.; Kong, D.; You, H.; Kong, F.; Tang, R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer 2020, 20, 1204. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, Z.-Y.; Gan, X.; Dai, H.; Cai, D.; Liu, R.-H.; Zhou, J.-M.; Zhang, H.-L.; Li, Z.-H.; Luo, Q.-Q.; et al. A novel four-gene signature for predicting the prognosis of hepatocellular carcinoma. Scand. J. Gastroenterol. 2022, 57, 1227–1237. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, T.; Liu, Z.; Xiao, C.; Du, J.; Zuo, S.; Li, H.; Gu, H. A Differentiation-Related Gene Prognostic Index Contributes to Prognosis and Immunotherapy Evaluation in Patients with Hepatocellular Carcinoma. Cells 2022, 11, 2302. [Google Scholar] [CrossRef]
- Wang, P.; Ouyang, L.; Zheng, L.; Wang, Z. Identifying hepatocellular carcinoma-related genes and pathways by system biology analysis. Ir. J. Med. Sci. 2015, 184, 357–364. [Google Scholar] [CrossRef]
- He, J.; Han, Z.; Liu, J.; Zhou, J.; Zou, M.; Lv, Y.; Li, Y.; Cao, M. Overexpression of Long Non-Coding RNA MEG3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells via Negative Modulation of miRNA-664. J. Cell. Biochem. 2017, 118, 3713–3721. [Google Scholar] [CrossRef] [PubMed]



| Patient ID | Sex | Age | State | Staining | Intensity | Quantity | More Details |
|---|---|---|---|---|---|---|---|
| 3402 | Female | 54 | Normal | Medium | Moderate | >75% | https://www.proteinatlas.org/ENSG00000198099-ADH4/tissue/liver#img (accessed on 5 October 2022) |
| 3222 | Female | 63 | Normal | Medium | Moderate | >75% | |
| 2429 | Male | 55 | Normal | Medium | Moderate | >75% | |
| 3215 | Female | 61 | HCC | Low | Weak | 25–75% | https://www.proteinatlas.org/ENSG00000198099-ADH4/pathology/liver+cancer#img (accessed on 5 October 2022) |
| 2766 | Female | 73 | HCC | Not detected | Negative | None | |
| 2177 | Female | 58 | HCC | Low | Weak | 25–75% | |
| 3196 | Male | 65 | HCC | Not detected | Negative | None | |
| 2280 | Male | 80 | HCC | Medium | Moderate | 25–75% | |
| 2556 | Male | 72 | HCC | Low | Weak | 25–75% | |
| 3346 | Female | 73 | HCC | Medium | Moderate | 25–75% | |
| 3477 | Male | 67 | HCC | Low | Weak | 25–75% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karunakara, S.H.; Puttahanumantharayappa, L.D.; Sannappa Gowda, N.G.; Shiragannavar, V.D.; Santhekadur, P.K. Novel Insights into MEG3/miR664a-3p/ADH4 Axis and Its Possible Role in Hepatocellular Carcinoma from an in Silico Perspective. Genes 2022, 13, 2254. https://doi.org/10.3390/genes13122254
Karunakara SH, Puttahanumantharayappa LD, Sannappa Gowda NG, Shiragannavar VD, Santhekadur PK. Novel Insights into MEG3/miR664a-3p/ADH4 Axis and Its Possible Role in Hepatocellular Carcinoma from an in Silico Perspective. Genes. 2022; 13(12):2254. https://doi.org/10.3390/genes13122254
Chicago/Turabian StyleKarunakara, Shreyas H., Lakshana D. Puttahanumantharayappa, Nirmala G. Sannappa Gowda, Varsha D. Shiragannavar, and Prasanna K. Santhekadur. 2022. "Novel Insights into MEG3/miR664a-3p/ADH4 Axis and Its Possible Role in Hepatocellular Carcinoma from an in Silico Perspective" Genes 13, no. 12: 2254. https://doi.org/10.3390/genes13122254
APA StyleKarunakara, S. H., Puttahanumantharayappa, L. D., Sannappa Gowda, N. G., Shiragannavar, V. D., & Santhekadur, P. K. (2022). Novel Insights into MEG3/miR664a-3p/ADH4 Axis and Its Possible Role in Hepatocellular Carcinoma from an in Silico Perspective. Genes, 13(12), 2254. https://doi.org/10.3390/genes13122254








