Rare CNVs and Known Genes Linked to Macrocephaly: Review of Genomic Loci and Promising Candidate Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. DECIPHER Patients
2.2. Literature Review of Known Macrocephaly Genes and Associated CNV Syndromes
3. Results
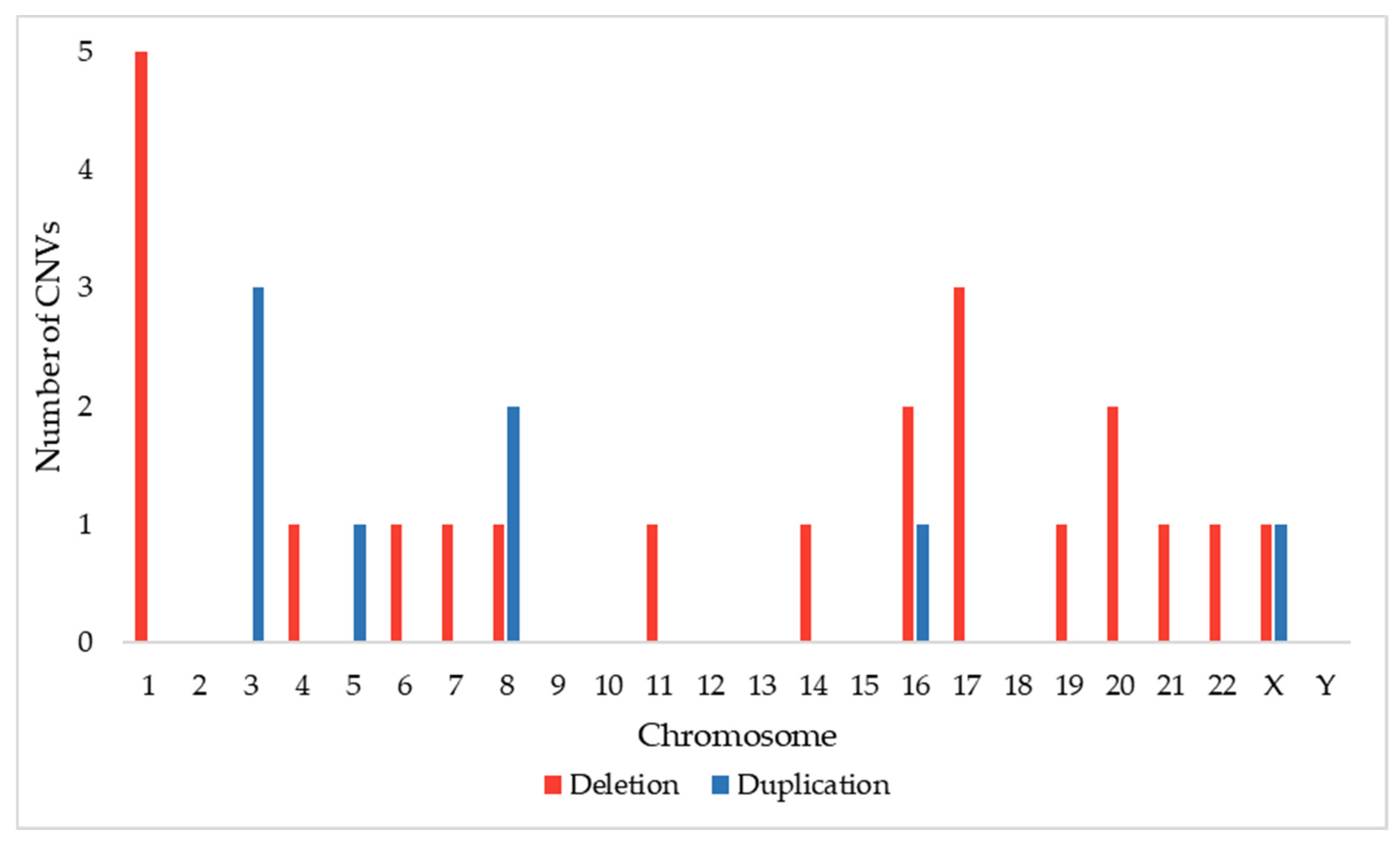
3.1. CNVs Associated with Macrocephaly in the DECIPHER Database
3.2. Literature Review of Macrocephaly Genes and Associated CNV Syndromes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Accogli, A.; Geraldo, A.F.; Piccolo, G.; Riva, A.; Scala, M.; Balagura, G.; Salpietro, V.; Madia, F.; Maghnie, M.; Zara, F.; et al. Diagnostic Approach to Macrocephaly in Children. Front. Pediatr. 2022, 9, 1534. [Google Scholar] [CrossRef] [PubMed]
- Olney, A.H. Macrocephaly Syndromes. Semin. Pediatr. Neurol. 2007, 14, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.P.; Mankad, K.; Gonçalves, F.G.; Talenti, G.; Alexia, E. Macrocephaly. Top. Magn. Reson. Imaging 2018, 27, 197–217. [Google Scholar] [CrossRef]
- Thomas, C.N.; Kolbe, A.B.; Binkovitz, L.A.; McDonald, J.S.; Thomas, K.B. Asymptomatic macrocephaly: To scan or not to scan. Pediatr. Radiol. 2021, 51, 811–821. [Google Scholar] [CrossRef]
- Williams, C.A.; Dagli, A.; Battaglia, A. Genetic disorders associated with macrocephaly. Am. J. Med. Genet. Part A 2008, 146A, 2023–2037. [Google Scholar] [CrossRef] [PubMed]
- Malinger, G.; Lev, D.; Ben-Sira, L.; Hoffmann, C.; Herrera, M.; Viñals, F.; Vinkler, H.; Ginath, S.; Biran-Gol, Y.; Kidron, D.; et al. Can syndromic macrocephaly be diagnosed in utero? Ultrasound Obstet. Gynecol. 2010, 37, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Orru, E.; Calloni, S.F.; Tekes, A.; Huisman, T.A.G.M.; Soares, B.P. The Child With Macrocephaly: Differential Diagnosis and Neuroimaging Findings. Am. J. Roentgenol. 2018, 210, 848–859. [Google Scholar] [CrossRef]
- Winden, K.D.; Yuskaitis, C.J.; Poduri, A. Megalencephaly and Macrocephaly. Skull Base 2015, 35, 277–287. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ross, A.H. Why is PTEN an important tumor suppressor? J. Cell. Biochem. 2007, 102, 1368–1374. [Google Scholar] [CrossRef]
- Hopkins, B.D.; Hodakoski, C.; Barrows, D.; Mense, S.M.; Parsons, R.E. PTEN function: The long and the short of it. Trends Biochem. Sci. 2014, 39, 183–190. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, W.-C.; Séjourné, J.; Clipperton-Allen, A.E.; Page, D.T. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated-Catenin Signaling. J. Neurosci. 2015, 35, 10252–10267. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Junco-Clemente, P.; Golshani, P. PTEN. Commun. Integr. Biol. 2014, 7, e28358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, M.G.; Dasouki, M.J.; Zhou, X.P.; Talebizadeh, Z.; Brown, M.; Takahashi, T.N.; Miles, J.H.; Wang, C.H.; Stratton, R.; Pilarski, R.; et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005, 42, 318–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B., Jr.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Prim. 2017, 3, 17065. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shin, J.; Risgaard, R.D.; Parries, M.J.; Wang, J.; Chasman, D.; Liu, S.; Roy, S.; Bhattacharyya, A.; Zhao, X. Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res. 2020, 30, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Pirozzi, F.; Nelson, B.; Mirzaa, G. From microcephaly to megalencephaly: Determinants of brain size. Dialog Clin. Neurosci. 2018, 20, 267–282. [Google Scholar] [CrossRef] [Green Version]
- Tatton-Brown, K.; Weksberg, R. Molecular Mechanisms of Childhood Overgrowth. Am. J. Med. Genet. Part C Semin. Med. Genet. 2013, 163, 71–75. [Google Scholar] [CrossRef]
- Wang, D.; Zeesman, S.; Tarnopolsky, M.A.; Nowaczyk, M.J. Duplication of AKT3 as a cause of macrocephaly in duplication 1q43q44. Am. J. Med. Genet. Part A 2013, 161, 2016–2019. [Google Scholar] [CrossRef]
- Lopes, F.; Torres, F.; Soares, G.; van Karnebeek, C.D.; Martins, C.; Antunes, D.; Silva, J.; Muttucomaroe, L.; Botelho, L.F.; Sousa, S.; et al. The Role of AKT3 Copy Number Changes in Brain Abnormalities and Neurodevelopmental Disorders: Four New Cases and Literature Review. Front. Genet. 2019, 10, 58. [Google Scholar] [CrossRef] [Green Version]
- Alcantara, D.; E Timms, A.; Gripp, K.; Baker, L.; Park, K.; Collins, S.; Cheng, C.; Stewart, F.; Mehta, S.G.; Saggar, A.; et al. Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain 2017, 140, 2610–2622. [Google Scholar] [CrossRef]
- Freeman, J.L.; Perry, G.H.; Feuk, L.; Redon, R.; McCarroll, S.A.; Altshuler, D.M.; Aburatani, H.; Jones, K.W.; Tyler-Smith, C.; Hurles, M.E.; et al. Copy number variation: New insights in genome diversity. Genome Res. 2006, 16, 949–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pös, O.; Radvanszky, J.; Styk, J.; Pös, Z.; Buglyó, G.; Kajsik, M.; Budis, J.; Nagy, B.; Szemes, T. Copy Number Variation: Methods and Clinical Applications. Appl. Sci. 2021, 11, 819. [Google Scholar] [CrossRef]
- Rice, A.M.; McLysaght, A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat. Commun. 2017, 8, 14366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, C.; Knijnenburg, J.; Bakker, B.; Vianna-Morgante, A.M.; Sloos, W.; A Otto, P.; Kriek, M.; Hansson, K.; Krepischi, A.; Fiegler, H.; et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: Clinical significance of imbalances present both in affected children and normal parents. J. Med. Genet. 2005, 43, 180–186. [Google Scholar] [CrossRef] [Green Version]
- Krepischi-Santos, A.; Vianna-Morgante, A.; Jehee, F.; Passos-Bueno, M.; Knijnenburg, J.; Szuhai, K.; Sloos, W.; Mazzeu, J.; Kok, F.; Cheroki, C.; et al. Whole-genome array-CGH screening in undiagnosed syndromic patients: Old syndromes revisited and new alterations. Cytogenet. Genome Res. 2006, 115, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.L.; Wain, K.E.; Oetjens, M.T.; Tolwinski, K.; Palen, E.; Hare-Harris, A.; Habegger, L.; Maxwell, E.K.; Reid, J.G.; Walsh, L.K.; et al. Identification of Neuropsychiatric Copy Number Variants in a Health Care System Population. JAMA Psychiatry 2020, 77, 1276. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.L.; Glessner, J.T.; Porcu, E.; Lepamets, M.; Brandon, R.; Lauricella, C.; Han, L.; Morley, T.; Niestroj, L.-M.; Ulirsch, J.; et al. A cross-disorder dosage sensitivity map of the human genome. Cell 2022, 185, 3041–3055.e25. [Google Scholar] [CrossRef]
- Cooper, G.M.; Coe, B.P.; Girirajan, S.; A Rosenfeld, J.; Vu, T.H.; Baker, C.; Williams, C.; Stalker, H.; Hamid, R.; Hannig, V.; et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011, 43, 838–846. [Google Scholar] [CrossRef] [Green Version]
- Firth, H.V.; Richards, S.M.; Bevan, A.P.; Clayton, S.; Corpas, M.; Rajan, D.; Van Vooren, S.; Moreau, Y.; Pettett, R.M.; Carter, N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009, 84, 524–533. [Google Scholar] [CrossRef] [Green Version]
- Abdin, D.; Rump, A.; Tzschach, A.; Sarnow, K.; Schröck, E.; Hackmann, K.; Di Donato, N. PUF60-SCRIB fusion transcript in a patient with 8q24.3 microdeletion and atypical Verheij syndrome. Eur. J. Med. Genet. 2018, 62, 103587. [Google Scholar] [CrossRef]
- Berecki, G.; Helbig, K.L.; Ware, T.L.; Grinton, B.; Skraban, C.M.; Marsh, E.D.; Berkovic, S.F.; Petrou, S. Novel Missense CACNA1G Mutations Associated with Infantile-Onset Developmental and Epileptic Encephalopathy. Int. J. Mol. Sci. 2020, 21, 6333. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, E.; Collins, A.; Papa, F.; Tejada, M.-I.; Wheeler, P.; Peeters, E.; Gijsbers, A.; van de Kamp, J.; Kriek, M.; Losekoot, M.; et al. Xq28 duplications including MECP2 in five females: Expanding the phenotype to severe mental retardation. Eur. J. Med. Genet. 2012, 55, 404–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunii, M.; Doi, H.; Hashiguchi, S.; Matsuishi, T.; Sakai, Y.; Iai, M.; Okubo, M.; Nakamura, H.; Takahashi, K.; Katsumoto, A.; et al. De novo CACNA1G variants in developmental delay and early-onset epileptic encephalopathies. J. Neurol. Sci. 2020, 416, 117047. [Google Scholar] [CrossRef] [PubMed]
- Santoro, C.; Malan, V.; Bertoli, M.; Boddaert, N.; Vidaud, D.; Lyonnet, S. Sporadic NF1 mutation associated with a de-novo 20q11.3 deletion explains the association of unusual facies, Moyamoya vasculopathy, and developmental delay, reported by Bertoli et al. in 2009. Clin. Dysmorphol. 2013, 22, 42–43. [Google Scholar] [CrossRef]
- Zahir, F.; Firth, H.V.; Baross, A.; Delaney, A.D.; Eydoux, P.; Gibson, W.; Langlois, S.; Martin, H.; Willatt, L.; A Marra, M.; et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J. Med. Genet. 2007, 44, 556–561. [Google Scholar] [CrossRef] [Green Version]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Clinical Genome Resource, Curations of Recurrent CNVs. Available online: https://search.clinicalgenome.org/kb/gene-dosage/cnv?page=1&size=All&search= (accessed on 17 November 2022).
- DECIPHER, CNV Syndromes. Available online: https://www.deciphergenomics.org/disorders/syndromes/list (accessed on 17 November 2022).
- Jayasena, C.S.; Bronner, M.E. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-β signaling. J. Cell Biol. 2012, 199, 453–466. [Google Scholar] [CrossRef] [Green Version]
- Koehler, U.; Holinski-Feder, E.; Ertl-Wagner, B.; Kunz, J.; Von Moers, A.; Von Voss, H.; Schell-Apacik, C. A novel 1p31.3p32.2 deletion involving the NFIA gene detected by array CGH in a patient with macrocephaly and hypoplasia of the corpus callosum. Eur. J. Pediatr. 2009, 169, 463–468. [Google Scholar] [CrossRef]
- Brunetti-Pierri, N.; Berg, J.; Scaglia, F.; Belmont, J.; A Bacino, C.; Sahoo, T.; Lalani, S.R.; Graham, B.; Lee, B.; Shinawi, M.; et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008, 40, 1466–1471. [Google Scholar] [CrossRef]
- Molin, A.-M.; Andrieux, J.; A Koolen, D.; Malan, V.; Carella, M.; Colleaux, L.; Cormier-Daire, V.; David, A.; De Leeuw, N.; Delobel, B.; et al. A novel microdeletion syndrome at 3q13.31 characterised by developmental delay, postnatal overgrowth, hypoplastic male genitals, and characteristic facial features. J. Med. Genet. 2011, 49, 104–109. [Google Scholar] [CrossRef]
- Vuillaume, M.-L.; Delrue, M.-A.; Naudion, S.; Toutain, J.; Fergelot, P.; Arveiler, B.; Lacombe, D.; Rooryck, C. Expanding the clinical phenotype at the 3q13.31 locus with a new case of microdeletion and first characterization of the reciprocal duplication. Mol. Genet. Metab. 2013, 110, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Cheung, S.-W.; Breman, A.M.; Bacino, C.A. 4p16.3 microdeletions and microduplications detected by chromosomal microarray analysis: New insights into mechanisms and critical regions. Am. J. Med. Genet. Part A 2016, 170, 2540–2550. [Google Scholar] [CrossRef] [PubMed]
- Weise, A.; Mrasek, K.; Klein, E.; Mulatinho, M.; Llerena, J.; Hardekopf, D.; Pekova, S.; Bhatt, S.; Kosyakova, N.; Liehr, T.; et al. Microdeletion and Microduplication Syndromes. J. Histochem. Cytochem. 2012, 60, 346–358. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Zhang, F.; Brundage, E.; Scheuerle, A.; Lanpher, B.; Erickson, R.P.; Powis, Z.; Robinson, H.B.; Trapane, P.L.; Stachiw-Hietpas, D.; et al. Genomic duplication resulting in increased copy number of genes encoding the sister chromatid cohesion complex conveys clinical consequences distinct from Cornelia de Lange. J. Med. Genet. 2008, 46, 626–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, L.M.; De Ravel, T.; Graham, B.H.; Frenkel, S.M.; Van Driessche, J.; Stankiewicz, P.; Lupski, J.R.; Vermeesch, J.R.; Cheung, S.W. A syndrome of short stature, microcephaly and speech delay is associated with duplications reciprocal to the common Sotos syndrome deletion. Eur. J. Hum. Genet. 2009, 18, 258–261. [Google Scholar] [CrossRef] [Green Version]
- Chui, J.V.; Weisfeld-Adams, J.D.; Tepperberg, J.; Mehta, L. Clinical and molecular characterization of chromosome 7p22.1 microduplication detected by array CGH. Am. J. Med. Genet. Part A 2011, 155, 2508–2511. [Google Scholar] [CrossRef]
- Bosfield, K.; Diaz, J.; Leon, E. Pure Distal 7q Duplication: Describing a Macrocephalic Neurodevelopmental Syndrome, Case Report and Review of the Literature. Mol. Syndr. 2021, 12, 159–168. [Google Scholar] [CrossRef]
- Akcakaya, N.H.; Capan, Y.; Schulz, H.; Sander, T.; Caglayan, S.H.; Yapıcı, Z. De novo 8p23.1 deletion in a patient with absence epilepsy. Epileptic Disord. 2017, 19, 217–221. [Google Scholar] [CrossRef]
- Ballarati, L.; Cereda, A.; Caselli, R.; Selicorni, A.; Recalcati, M.P.; Maitz, S.; Finelli, P.; Larizza, L.; Giardino, D. Genotype–phenotype correlations in a new case of 8p23.1 deletion and review of the literature. Eur. J. Med. Genet. 2010, 54, 55–59. [Google Scholar] [CrossRef]
- Vargiami, E.; Ververi, A.; Kyriazi, M.; Papathanasiou, E.; Gioula, G.; Gerou, S.; Al-Mutawa, H.; Kambouris, M.; Zafeiriou, D.I. Severe clinical presentation in monozygotic twins with 10p15.3 microdeletion syndrome. Am. J. Med. Genet. Part A 2013, 164, 764–768. [Google Scholar] [CrossRef]
- DeScipio, C.; Conlin, L.; Rosenfeld, J.; Tepperberg, J.; Pasion, R.; Patel, A.; McDonald, M.T.; Aradhya, S.; Ho, D.; Goldstein, J.; et al. Subtelomeric deletion of chromosome 10p15.3: Clinical findings and molecular cytogenetic characterization. Am. J. Med. Genet. Part A 2012, 158A, 2152–2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumiene, B.; Čiuladaitė, Ž.; Preikšaitienė, E.; Mameniškienė, R.; Utkus, A.; Kučinskas, V. Phenotype comparison confirms ZMYND11 as a critical gene for 10p15.3 microdeletion syndrome. J. Appl. Genet. 2017, 58, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Siavrienė, E.; Preikšaitienė, E.; Maldžienė, Ž.; Mikštienė, V.; Rančelis, T.; Ambrozaitytė, L.; Gueneau, L.; Reymond, A.; Kučinskas, V. A de novo 13q31.3 microduplication encompassing the miR-17 ~ 92 cluster results in features mirroring those associated with Feingold syndrome 2. Gene 2020, 753, 144816. [Google Scholar] [CrossRef] [PubMed]
- Yasin, H.; Gibson, W.; Langlois, S.; Stowe, R.M.; Tsang, E.; Lee, L.; Poon, J.; Tran, G.; Tyson, C.; Wong, C.K.; et al. A distinct neurodevelopmental syndrome with intellectual disability, autism spectrum disorder, characteristic facies, and macrocephaly is caused by defects in CHD8. J. Hum. Genet. 2019, 64, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Kriek, M.; Walenkamp, M.; Hansson, K.; van Rhijn, A.; Clayton-Smith, J.; Wit, J.; Breuning, M. Tall stature and duplication of the insulin-like growth factor I receptor gene. Eur. J. Med. Genet. 2007, 50, 1–10. [Google Scholar] [CrossRef]
- Qureshi, A.Y.; Mueller, S.; Snyder, A.Z.; Mukherjee, P.; Berman, J.I.; Roberts, T.P.; Nagarajan, S.S.; Spiro, J.E.; Chung, W.K.; Sherr, E.H.; et al. Opposing Brain Differences in 16p11.2 Deletion and Duplication Carriers. J. Neurosci. 2014, 34, 11199–11211. [Google Scholar] [CrossRef] [Green Version]
- Mooneyham, K.A.; Holden, K.R.; Cathey, S.; Dwivedi, A.; Dupont, B.R.; Lyons, M.J. Neurodevelopmental delays and macrocephaly in 17p13.1 microduplication syndrome. Am. J. Med. Genet. Part A 2014, 164, 2887–2891. [Google Scholar] [CrossRef]
- Mefford, H.; Mitchell, E.; Hodge, J. 17q12 Recurrent Duplication. [Updated 13 January 2022]; In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2016; 1993–2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK344340/ (accessed on 27 June 2022).
- Zollino, M.; Marangi, G.; Ponzi, E.; Orteschi, D.; Ricciardi, S.; Lattante, S.; Murdolo, M.; Battaglia, D.; Contaldo, I.; Mercuri, E.; et al. Intragenic KANSL1 mutations and chromosome 17q21.31 deletions: Broadening the clinical spectrum and genotype–phenotype correlations in a large cohort of patients. J. Med. Genet. 2015, 52, 804–814. [Google Scholar] [CrossRef]
- Nevado, J.; Rosenfeld, J.A.; Mena, R.; Palomares-Bralo, M.; Vallespin, E.; Angeles Mori, M.; Tenorio, J.A.; Gripp, K.W.; Denenberg, E.; Del Campo, M.; et al. PIAS4 is associated with macro/microcephaly in the novel interstitial 19p13. 3 microdele-tion/microduplication syndrome. Eur. J. Hum. Genet. 2015, 23, 1615–1626. [Google Scholar] [CrossRef] [Green Version]
- Orellana, C.; Roselló, M.; Monfort, S.; Mayo, S.; Oltra, S.; Martínez, F. Pure duplication of 19p13.3 in three members of a family with intellectual disability and literature review. Definition of a new microduplication syndrome. Am. J. Med. Genet. Part A 2015, 167, 1614–1620. [Google Scholar] [CrossRef]
- Pinchefsky, E.; Laneuville, L.; Srour, M. Distal 22q11.2 Microduplication. Child Neurol. Open 2017, 4, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buzanello, A.; Ramos, A.C.; Silva, A.P.; Ceron, C.; Schmitt, J.; Menegais, K.; Ribeiro, N.D.; Siepmann, R.; Nardi, A. Encéfalo: Estruturas e Funções. Ação Odonto 2013, 1, 10. [Google Scholar]
- Deshpande, A.; Weiss, L.A. Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Dev. Neurobiol. 2018, 78, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Pös, O.; Radvanszky, J.; Buglyó, G.; Pös, Z.; Rusnakova, D.; Nagy, B.; Szemes, T. DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects. Biomed. J. 2021, 44, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Golzio, C.; Katsanis, N. Genetic architecture of reciprocal CNVs. Curr. Opin. Genet. Dev. 2013, 23, 240–248. [Google Scholar] [CrossRef] [Green Version]
- Zarrei, M.; MacDonald, J.; Merico, D.; Scherer, S. A copy number variation map of the human genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef]
- Tolezano, G.; Bastos, G.; Krepischi, A. Burden of rare copy number variants in microcephaly: A Brazilian cohort of 185 microcephalic patients and review of the literature. J. Autism Dev. Disord. 2022; accepted. [Google Scholar]
- Dikow, N.; Maas, B.; Gaspar, H.; Kreiss-Nachtsheim, M.; Engels, H.; Kuechler, A.; Garbes, L.; Netzer, C.; Neuhann, T.M.; Koehler, U.; et al. The phenotypic spectrum of duplication 5q35.2-q35.3 encompassing NSD1: Is it really a reversed Sotos syndrome? Am. J. Med. Genet. Part A 2013, 161, 2158–2166. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Szybowska, M. Novel features of Helsmoortel-Van der Aa/ADNP syndrome in a boy with a known pathogenic mutation in the ADNP gene detected by exome sequencing. Am. J. Med. Genet. Part A 2017, 173, 1994–1995. [Google Scholar] [CrossRef]
- Gozes, I. The ADNP Syndrome and CP201 (NAP) Potential and Hope. Front. Neurol. 2020, 11, 608444. [Google Scholar] [CrossRef]
- Pinhasov, A.; Mandel, S.; Torchinsky, A.; Giladi, E.; Pittel, Z.; Goldsweig, A.M.; Servoss, S.J.; E Brenneman, D.; Gozes, I. Activity-dependent neuroprotective protein: A novel gene essential for brain formation. Dev. Brain Res. 2003, 144, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Peng, X.; Cao, Y.; Zhou, Y.; Sun, Y. ADNP promotes neural differentiation by modulating Wnt/β-catenin signaling. Nat. Commun. 2020, 11, 2984. [Google Scholar] [CrossRef] [PubMed]
- Revah-Politi, A.; Ganapathi, M.; Bier, L.; Cho, M.T.; Goldstein, D.B.; Hemati, P.; Iglesias, A.; Juusola, J.; Pappas, J.; Petrovski, S.; et al. Loss-of-function variants in NFIA provide further support that NFIA is a critical gene in 1p32-p31 deletion syndrome: A four patient series. Am. J. Med. Genet. Part A 2017, 173, 3158–3164. [Google Scholar] [CrossRef] [PubMed]
- Sieff, C. Diamond-Blackfan Anemia. [updated 17 June 2021]; In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2009; 1993–2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK7047/ (accessed on 11 July 2022).
- Chakraborty, A.; Uechi, T.; Nakajima, Y.; Gazda, H.T.; O’Donohue, M.-F.; Gleizes, P.-E.; Kenmochi, N. Cross talk between TP53 and c-Myc in the pathophysiology of Diamond-Blackfan anemia: Evidence from RPL11-deficient in vivo and in vitro models. Biochem. Biophys. Res. Commun. 2018, 495, 1839–1845. [Google Scholar] [CrossRef]
- Farrar, J.E.; Dahl, N. Untangling the Phenotypic Heterogeneity of Diamond Blackfan Anemia. Semin. Hematol. 2011, 48, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Mochida, G.H.; Mahajnah, M.; Hill, A.D.; Basel-Vanagaite, L.; Gleason, D.; Hill, R.S.; Bodell, A.; Crosier, M.; Straussberg, R.; Walsh, C.A. A Truncating Mutation of TRAPPC9 Is Associated with Autosomal-Recessive Intellectual Disability and Postnatal Microcephaly. Am. J. Hum. Genet. 2009, 85, 897–902. [Google Scholar] [CrossRef] [Green Version]
- Mbimba, T.; Hussein, N.J.; Najeed, A.; Safadi, F.F. TRAPPC9: Novel insights into its trafficking and signaling pathways in health and disease (Review). Int. J. Mol. Med. 2018, 42, 2991–2997. [Google Scholar] [CrossRef] [Green Version]
- Rasika, S.; Passemard, S.; Verloes, A.; Gressens, P.; El Ghouzzi, V. Golgipathies in Neurodevelopment: A New View of Old Defects. Dev. Neurosci. 2018, 40, 396–416. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.S.; Cimino, I.; Yalcin, B.; Raghupathy, N.; Vancollie, V.E.; Ibarra-Soria, X.; Firth, H.V.; Rimmington, D.; Farooqi, I.S.; Lelliott, C.J.; et al. Trappc9 deficiency causes parent-of-origin dependent microcephaly and obesity. PLoS Genet. 2020, 16, e1008916. [Google Scholar] [CrossRef]
- Personnic, N.; Lakisic, G.; Gouin, E.; Rousseau, A.; Gautreau, A.; Cossart, P.; Bierne, H. A role for Ral GTPase-activating protein subunit β in mitotic regulation. FEBS J. 2014, 281, 2977–2989. [Google Scholar] [CrossRef]
- Wagner, M.; Skorobogatko, Y.; Pode-Shakked, B.; Powell, C.M.; Alhaddad, B.; Seibt, A.; Barel, O.; Heimer, G.; Hoffmann, C.; Demmer, L.A.; et al. Bi-allelic Variants in RALGAPA1 Cause Profound Neurodevelopmental Disability, Muscular Hypotonia, Infantile Spasms, and Feeding Abnormalities. Am. J. Hum. Genet. 2020, 106, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.D.; Chen, X.-W.; Kaplan, R.; Saltiel, A.; Walker, C.L.; Reiner, D.J.; Der, C.J. Ral and Rheb GTPase Activating Proteins Integrate mTOR and GTPase Signaling in Aging, Autophagy, and Tumor Cell Invasion. Mol. Cell 2014, 53, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Clinical Genome Resource. RALGAPB Dosage Sensitivity. Available online: https://search.clinicalgenome.org/kb/gene-dosage/RALGAPB#report_details_haploinsufficiency (accessed on 22 November 2022).
- Cloëtta, D.; Thomanetz, V.; Baranek, C.; Lustenberger, R.M.; Lin, S.; Oliveri, F.; Atanasoski, S.; Rüegg, M.A. Inactivation of mTORC1 in the Developing Brain Causes Microcephaly and Affects Gliogenesis. J. Neurosci. 2013, 33, 7799–7810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Kwong, D.L.-W.; Zhu, C.-L.; Chen, L.-L.; Dong, S.-S.; Zhang, L.-Y.; Tian, J.; Qi, C.-B.; Cao, T.-T.; Wong, A.M.G.; et al. RBMS3 at 3p24 Inhibits Nasopharyngeal Carcinoma Development via Inhibiting Cell Proliferation, Angiogenesis, and Inducing Apoptosis. PLOS ONE 2012, 7, e44636. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wu, Y.; Liu, Y.; Pan, F.; Zeng, H.; Li, X.; Yu, L. Tumor Suppressor Effect of RBMS3 in Breast Cancer. Technol. Cancer Res. Treat. 2021, 20, 1–9. [Google Scholar] [CrossRef]
- Oo, H.Z.; Sentani, K.; Sakamoto, N.; Anami, K.; Naito, Y.; Uraoka, N.; Oshima, T.; Yanagihara, K.; Oue, N.; Yasui, W. Overexpression of ZDHHC14 promotes migration and invasion of scirrhous type gastric cancer. Oncol. Rep. 2014, 32, 403–410. [Google Scholar] [CrossRef] [Green Version]
- Yeste-Velasco, M.; Mao, X.; Grose, R.; Kudahetti, S.C.; Lin, D.; Marzec, J.; Vasiljević, N.; Chaplin, T.; Xue, L.; Xu, M.; et al. Identification of ZDHHC14 as a novel human tumour suppressor gene. J. Pathol. 2014, 232, 566–577. [Google Scholar] [CrossRef] [Green Version]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]

| DECIPHER ID | Chromosomal Microarray Analysis (CMA) | Genotype-Phenotype Correlation | References | ||||
|---|---|---|---|---|---|---|---|
| Cytoband | Genomic Coordinates (GRCh38) | CNV Type | Classification | Size (kb) | Included Genes | ||
| (Protein Coding) | |||||||
| 281752 | 1q21.3 | 1:153818614-153823418 | Deletion P | Pathogenic | 5 | GATAD2B | |
| 269967 | 1p36.11 | 1:23691668-23696616 | Deletion P | Pathogenic | 5 | RPL11 ↓ | |
| 358646 | 1p31.3 | 1:61091785-61304381 | Deletion P | Pathogenic | 213 | NFIA | |
| 288170 | 1p31.3 | 1:61151026-61379931 | Deletion VUS | Pathogenic | 229 | NFIA | |
| 360889 | 1p36.33 | 1:917483-1282652 | Deletion P | VUS | 365 | AGRN/B3GALT6/C1QTNF12/C1orf159/HES4/ISG15/KLHL17/NOC2L/PERM1/PLEKHN1/RNF223/SAMD11/SCNN1D/SDF4/TNFRSF18/TNFRSF4/TTLL10/UBE2J2 | |
| 249115 | 3q25.33 | 3:159979210-160186591 | Duplication U | Likely benign | 207 | IL12A | [32] |
| Xq28 | X:154032185-154347882 | Duplication U | Pathogenic | 316 | MECP2 ↓/OPN1LW/OPN1MW/OPN1MW2/OPN1MW3/TEX28/TKTL1 | ||
| 1376 | 3q27.2 | 3:185411752-185567638 | Duplication U | VUS | 156 | LIPH/MAP3K13/TMEM41A | |
| 370137 | 3p24.1 | 3:29248723-29450441 | Duplication LP | VUS | 202 | RBMS3 | |
| 289221 | 4q22.2 | 4:93137138-93292777 | Deletion VUS | VUS | 156 | GRID2 | |
| 267309 | 5q35.3 | 5:177176839-177225635 | Duplication U | VUS | 49 | NSD1 ↓ | |
| 376373 | 6q25.3 | 6:157361157-157681369 | Deletion VUS | VUS | 320 | ZDHHC14 | |
| 288535 | 7p15.3 | 7:20953880-21052118 | Deletion VUS | Likely benign | 98 | - | |
| 339955 | 8q24.3 | 8:140361494-140527307 | Duplication VUS | VUS | 166 | AGO2/CHRAC1/TRAPPC9 ↓ | |
| 371384 | 8q24.3 | 8:143809435-143822571 | Deletion P | Pathogenic | 13 | PUF60 ↓/SCRIB ↓ | [30] |
| 314265 | 8p23.1 | 8:6518891-6801141 | Duplication P | VUS | 282 | MCPH1 ↓/ANGPT2/AGPAT5 | |
| 251808 | 11q13.2 | 11:68120554-68519565 | Deletion U | Pathogenic | 399 | LRP5/KMT5B/CHKA/C11ORF24/PPP6R3 | |
| 976 | 14q11.2 | 14:21309552-21460179 | Deletion U | Pathogenic | 151 | CHD8/RAB2B/RPGRIP1/SUPT16H | [35] |
| 253656 | 16p11.2 | 16:29837876-30179218 | Deletion U | Pathogenic | 341 | TAOK2/TLCD3B/PPP4C/CDIPT/SEZ6L2/YPEL3/MAPK3/C16orf92/INO80E/TBX6/DOC2A/ALDOA/ASPHD1/KCTD13 ↓/TMEM219/HIRIP3/MVP/GDPD3 | |
| 285342 | 16p13.2 | 16:9732634-9770161 | Deletion LP | Pathogenic | 38 | GRIN2A ↓ | |
| 301907 | 17q11.2 | 17:31334752-31340757 | Deletion P | Pathogenic | 6 | NF1 ↓ | |
| 287501 | 17q21.33 | 17:50600602-50604425 | Deletion U | Likely benign | 4 | CACNA1G ↓ | [31,33] |
| 368685 | 17q25.3 | 17:82821407-83086677 | Deletion VUS | Likely benign | 265 | ZNF750/METRNL/TBCD ↓/B3GNTL1 | |
| 300874 | 19p13.3 | 19:3979570-4131262 | Deletion P | Pathogenic | 152 | EEF2/PIAS4/MAP2K2/ZBTB7A | |
| 270687 | 20q11.23 | 20:38565558-38765539 | Deletion U | VUS | 200 | RALGAPB/SLC32A1/ADIG/ARHGAP40/ACTR5 | [34] |
| 412759 | 20q13.13 | 20:50891372-50893349 | Deletion P | Pathogenic | 2 | ADNP ↓ | |
| 249393 | 21q22.11 | 21:33581937-33883538 | Deletion U | VUS | 302 | ITSN1↓/DONSON ↓/CRYZL1 | |
| 259449 | 22q13.2 | 22:41743074-42171084 | Deletion VUS | Pathogenic | 428 | CCDC134/CENPM/CYP2D6/CYP2D7/MEI1/NAGA/NDUFA6/PHETA2/SEPTIN3/SHISA8/SMDT1/SREBF2/TCF20/TNFRSF13C/WBP2NL | |
| 270868 | Xq26.2 | X:133552850-134042983 | Deletion U | Pathogenic | 490 | GPC3 | |
| 16p11.2 | 16:29552664-30095687 | Duplication P | 543 | ALDOA/ASPHD1/C16orf54/C16orf92/CDIPT/DOC2A/HIRIP3/INO80E/KCTD13 ↓/KIF22/MAS/MVP/PAGR1/PPP4C/PRRT2/QPRT/SEZ6L2/SPN/TAOK2/TBX6/TLCD3B/TMEM219/YPEL3/ZG16 | |||
| Gene | Function * | Brain Expression ꜝ | Type of Variant (Probable Effect) | Reference |
|---|---|---|---|---|
| TRAPPC9 ↓ | May function in neuronal cells differentiation. LoF associated with microcephaly r | Yes | Partial duplication (unknown) | (OMIM #613192) |
| RALGAPB | RALGAPB plays an essential role in mitosis by controlling the spatial and temporal activation of RAL GTPases in the spindle assembly checkpoint (SAC) and cytokinesis | Yes | Partial deletion (unknown) | (OMIM *618833) |
| RBMS3 | Rbms3 was shown to exhibit tumor suppressor function via regulation of c-Myc and to bind/stabilize RNA in vitro. In zebrafish, LoF disrupts craniofacial development; not previously related to human diseases | Yes | Intragenic duplication (LoF) | (OMIM *300027) [39] |
| ZDHHC14 | Overexpression of ZDHHC14 reduces cell viability and induces apoptosis by activating a classic caspase-dependent pathway, whereas heterozygous knockout of ZDHHC14 increased colony formation ability of cells. | Yes | Entire deletion (LoF) | (OMIM *619295) |
| Condition | OMIM # | Genomic Coordinates (hg38) | Inheritance Pattern | Association with Head Size | ClinGen/ DECIPHER | Additional References |
|---|---|---|---|---|---|---|
| Chromosome 1p32-p31 deletion syndrome | 613735 | chr1:58193565-63125273 | Incomplete penetrance | Macrocephaly | … | [40] |
| Chromosome 1q21.1 duplication syndrome | 612475 | chr1:147105904-147917509 | Incomplete penetrance | Macrocephaly in duplication/Microcephaly in deletion (#612474) | Yes | [41] |
| Chromosome 2q31.2 deletion syndrome | 612345 | chr2:177100000-179700000 | Incomplete penetrance | Macrocephaly/Microcephaly | ... | - |
| Chromosome 3q13.31 deletion syndrome | 615433 | chr3:113700000-117600000 | de novo | Macrocephaly in deletion/Normal OFC in duplication | ... | [42,43] |
| Chromosome 4pter duplication syndrome | N/A | chr4:337779-2009235 | de novo | Macrocephaly in duplication/Microcephaly in deletion | Yes | [44] |
| Chromosome 4q32.1-q32.2 triplication syndrome | 613603 | chr4:154600000-163600000 | de novo | Macrocephaly | ... | [45] |
| Chromosome 5p13 duplication syndrome | 613174 | chr5:36845462–37231819 | de novo | Macrocephaly | ... | [46] |
| Chromosome 5q35 deletion syndrome (Sotos 1) | 117550 | chr5:176297633-177625115 | de novo | Macrocephaly in deletion/Microcephaly in duplication | Yes | [47] |
| Chromosome 7p22.1 duplication syndrome | N/A | chr7:5527147-5530600 | de novo | Relative macrocephaly in duplication/Microcephaly in deletion | ... | [48] |
| Chromosome 7q11.23 duplication syndrome | 609757 | chr7: 73330452-74728172 | Mostly de novo | Macrocephaly in duplication/Microcephaly in deletion (#194050) | Yes | - |
| Chromosome distal 7q (7(q32→qter)) duplication syndrome | N/A | chr7:128308047– 159119707 | Incomplete penetrance (can be inherited) | Macrocephaly | Yes ! | [49] |
| Chromosome 8p23.1 duplication syndrome | N/A | chr8:8242542-11908820 | de novo | Macrocephaly in duplication/Microcephaly in deletion | Yes | [28,50,51] |
| Chromosome 10p15.3 deletion syndrome | N/A | chr10:171237-2880776 | Incomplete penetrance (can be inherited) | Macrocephaly/Microcephaly | ... | [52,53,54] |
| Chromosome 10q22.3-q23.2 deletion syndrome | 612242 | chr10:80300000-95300000 | de novo | Macrocephaly in deletion/Microcephaly in duplication | Yes | - |
| Chromosome 11q deletion syndrome (Jacobsen) | 147791 | chr11:114600000-135086622 | de novo | Macrocephaly/Microcephaly | Yes | - |
| Chromosome 13q31.3 microduplication syndrome | N/A | chr13:91337007-91852603 | de novo | Macrocephaly in duplication/Microcephaly in deletion | ... | [55] |
| Chromosome 14q11.2 microdeletion syndrome | N/A | chr14:21359783-21393052 | de novo | Macrocephaly | … | [56] |
| Chromosome 15q11q13 deletion (PWS) | 176270 | chr15:22832519-28379874 | de novo | Macro-microcephaly in PWS/Microcephaly in AS | Yes | - |
| Chromosome 15q26qter duplication syndrome | 612626 | chr15:88500000-101991189 | de novo | Macrocephaly in duplication/Microcephaly in deletion | Yes | [57] |
| Chromosome 16p11.2 deletion syndrome, 593kb | 611913 | chr16:29595531-30188534 | Incomplete penetrance (can be inherited) | Macrocephaly in deletion/Microcephaly in duplication | Yes | [58] |
| Chromosome 17p13.1 duplication syndrome | N/A | chr17:7584958-8092957 | de novo | Macrocephaly in duplication/Microcephaly in deletion | ... | [45,59] |
| Chromosome 17q11.2 recurrent region 1.4Mb (del/dup) | 613675 618874 | chr17:30780079-31937008 | Del-de novo; Dup-incomplete penetrance (can be inherited) | Macrocephaly/Microcephaly | Yes | - |
| Chromosome 17q12 deletion syndrome | 614527 | chr17:36458167-37854616 | Mostly de novo | Macrocephaly in deletion/Microcephaly in duplication | Yes | [60] |
| Chromosome 17q21.31 deletion syndrome | 610443 | chr17:45627800-46087514 | Description of inherited cases | Macrocephaly in deletion/Microcephaly in duplication (#613533) | Yes | [61] |
| Chromosome 19p13.13 deletion syndrome | 613638 | chr19:12821186-13132186 | de novo | Macrocephaly in deletion (#613638)/Microcephaly in duplication | ... | [45] |
| Chromosome 19p13.3 microdeletion syndrome | N/A | chr19:2329321-4996917 | Description of inherited cases, although the majority are de novo | Macrocephaly in deletion/Microcephaly in duplication | … | [62,63] |
| Chromosome 22q11.2 duplication syndrome, distal | N/A | chr22:21562828-23306924 | Incomplete penetrance (can be inherited) | Macro-microcephaly in duplication/Microcephaly in deletion | Yes | [64] |
| Chromosome Xq22.3 telomeric deletion syndrome (AMME) | 300194 | chrX:104500000-109400000 | Dominant X-linked (description of inherited cases) | Macrocephaly | ... | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastos, G.C.; Tolezano, G.C.; Krepischi, A.C.V. Rare CNVs and Known Genes Linked to Macrocephaly: Review of Genomic Loci and Promising Candidate Genes. Genes 2022, 13, 2285. https://doi.org/10.3390/genes13122285
Bastos GC, Tolezano GC, Krepischi ACV. Rare CNVs and Known Genes Linked to Macrocephaly: Review of Genomic Loci and Promising Candidate Genes. Genes. 2022; 13(12):2285. https://doi.org/10.3390/genes13122285
Chicago/Turabian StyleBastos, Giovanna Civitate, Giovanna Cantini Tolezano, and Ana Cristina Victorino Krepischi. 2022. "Rare CNVs and Known Genes Linked to Macrocephaly: Review of Genomic Loci and Promising Candidate Genes" Genes 13, no. 12: 2285. https://doi.org/10.3390/genes13122285
APA StyleBastos, G. C., Tolezano, G. C., & Krepischi, A. C. V. (2022). Rare CNVs and Known Genes Linked to Macrocephaly: Review of Genomic Loci and Promising Candidate Genes. Genes, 13(12), 2285. https://doi.org/10.3390/genes13122285






