Identification of Hydroxylation Enzymes and the Metabolic Analysis of Dihydromyricetin Synthesis in Ampelopsis grossedentata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Compounds, Standards and Plant Samples
2.2. Genes, Plasmids and Strains
2.3. Plant Cultivation and RNA-Seq
2.4. Genes Amplification and Analyzation
2.5. Strains Growth Media and Culture Conditions
2.6. DHM Extraction from A. grossedentata
2.7. Quantitative HPLC Analysis
2.8. Data Processing and Statistical Analysis
3. Results
3.1. Spatial DHM Content in A. grossedentata
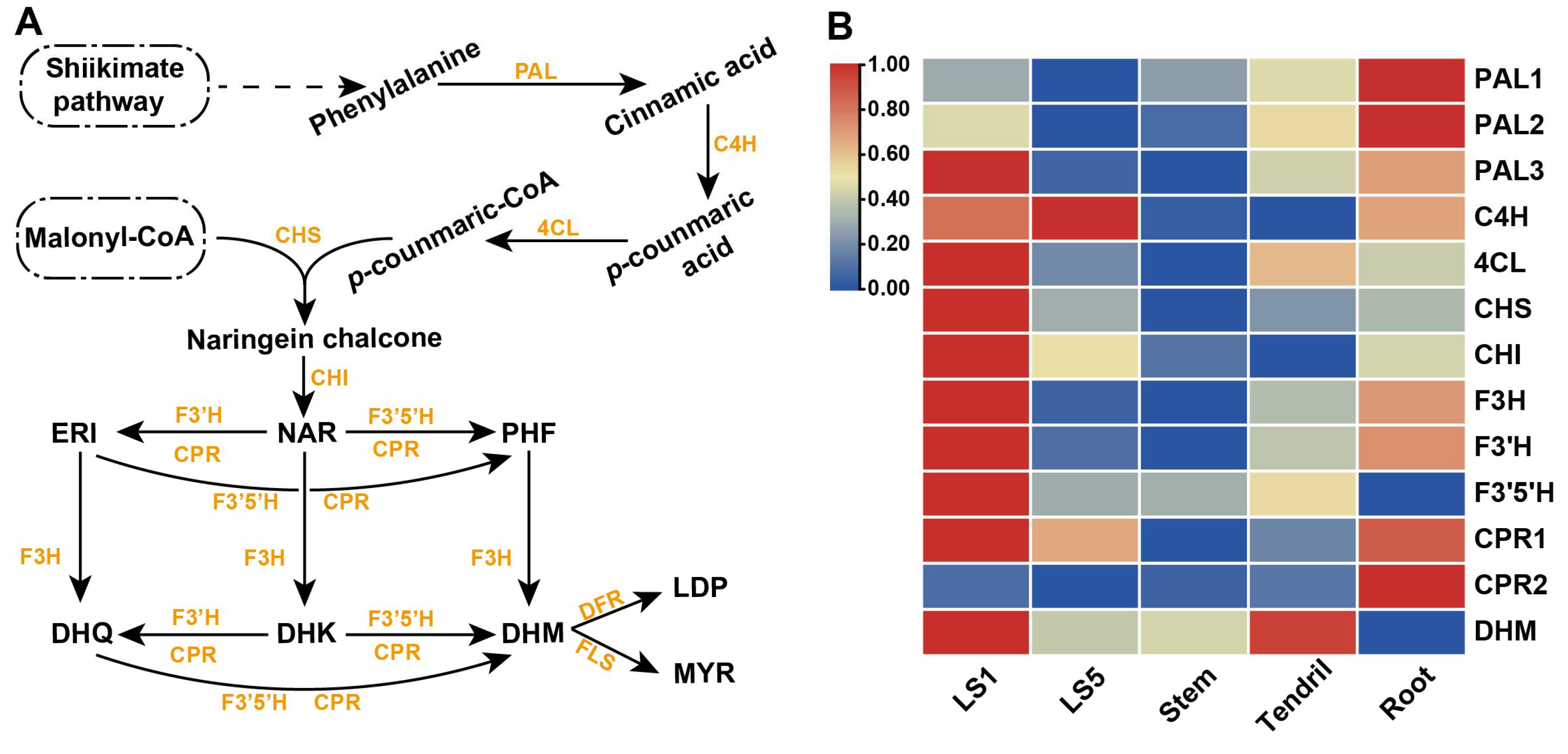
3.2. Analysis of Dihydroflavonol Metabolic Pathway and Genes Expression in A. grossedentata
3.3. Amplification and Analysis of Key Genes Related to DHM Biosynthesis
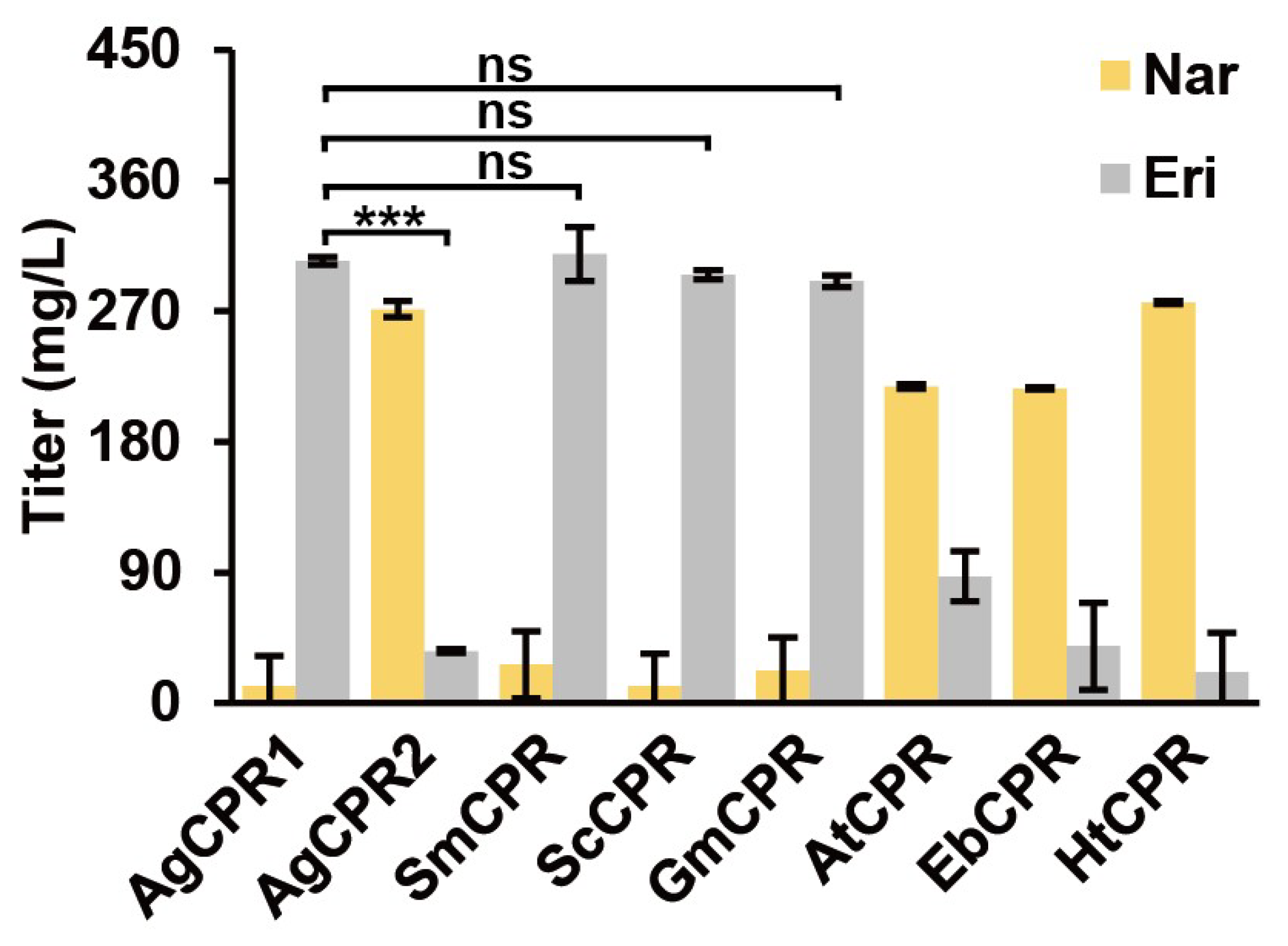
3.4. Comparison of AgCPRs and CPRs from Previous Studies
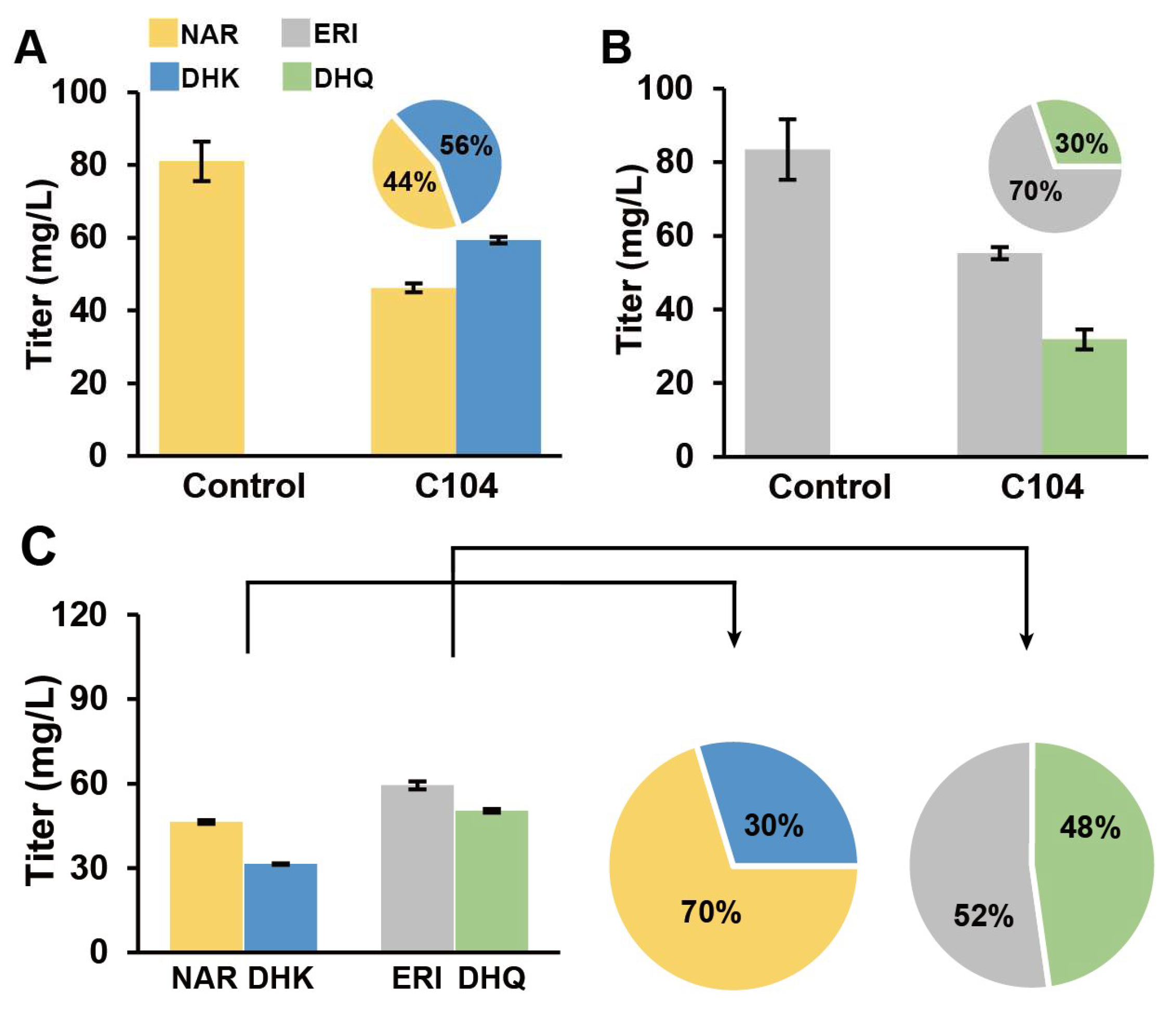
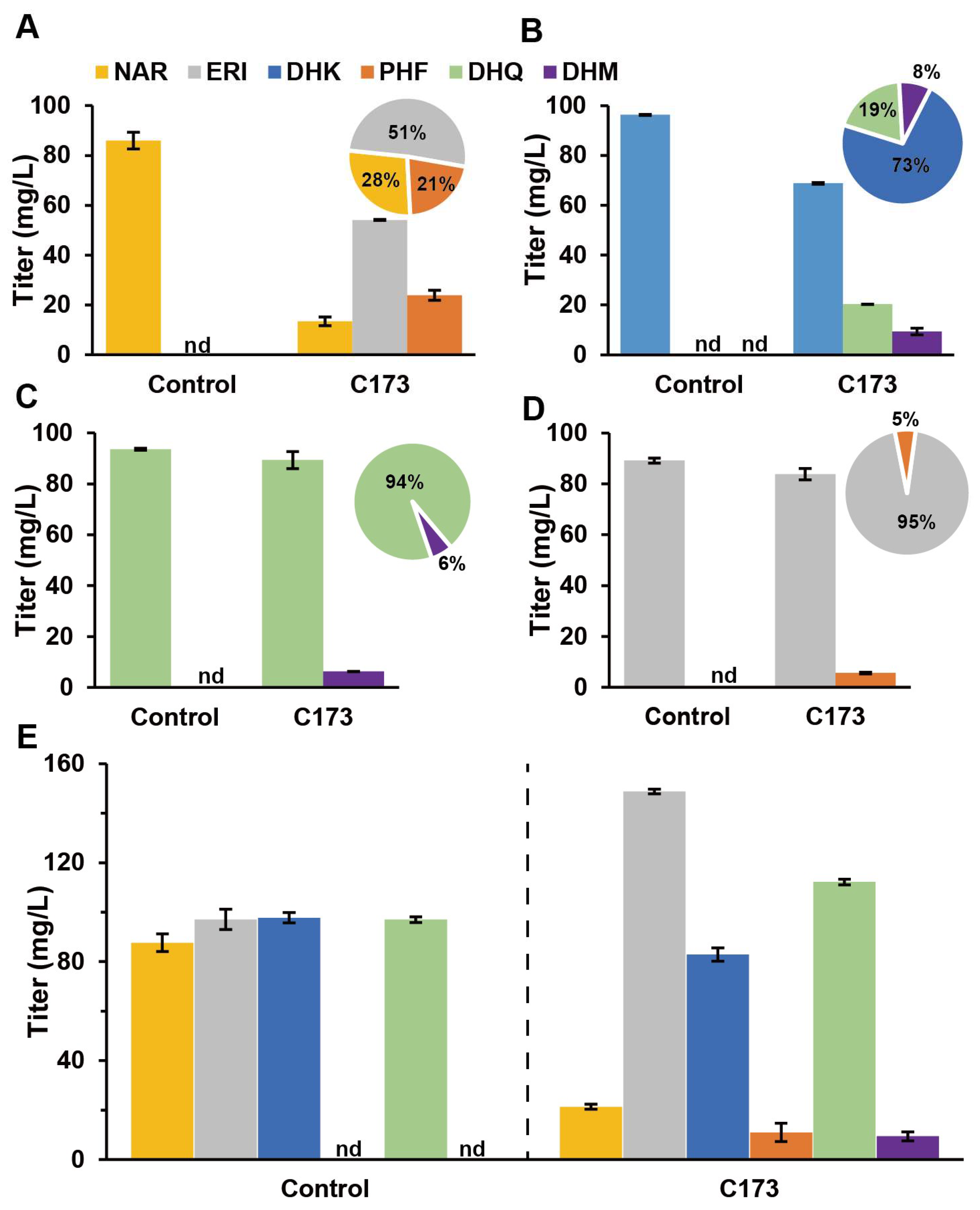
3.5. AgF3H and AgF3′H Functional and Efficiency Validation
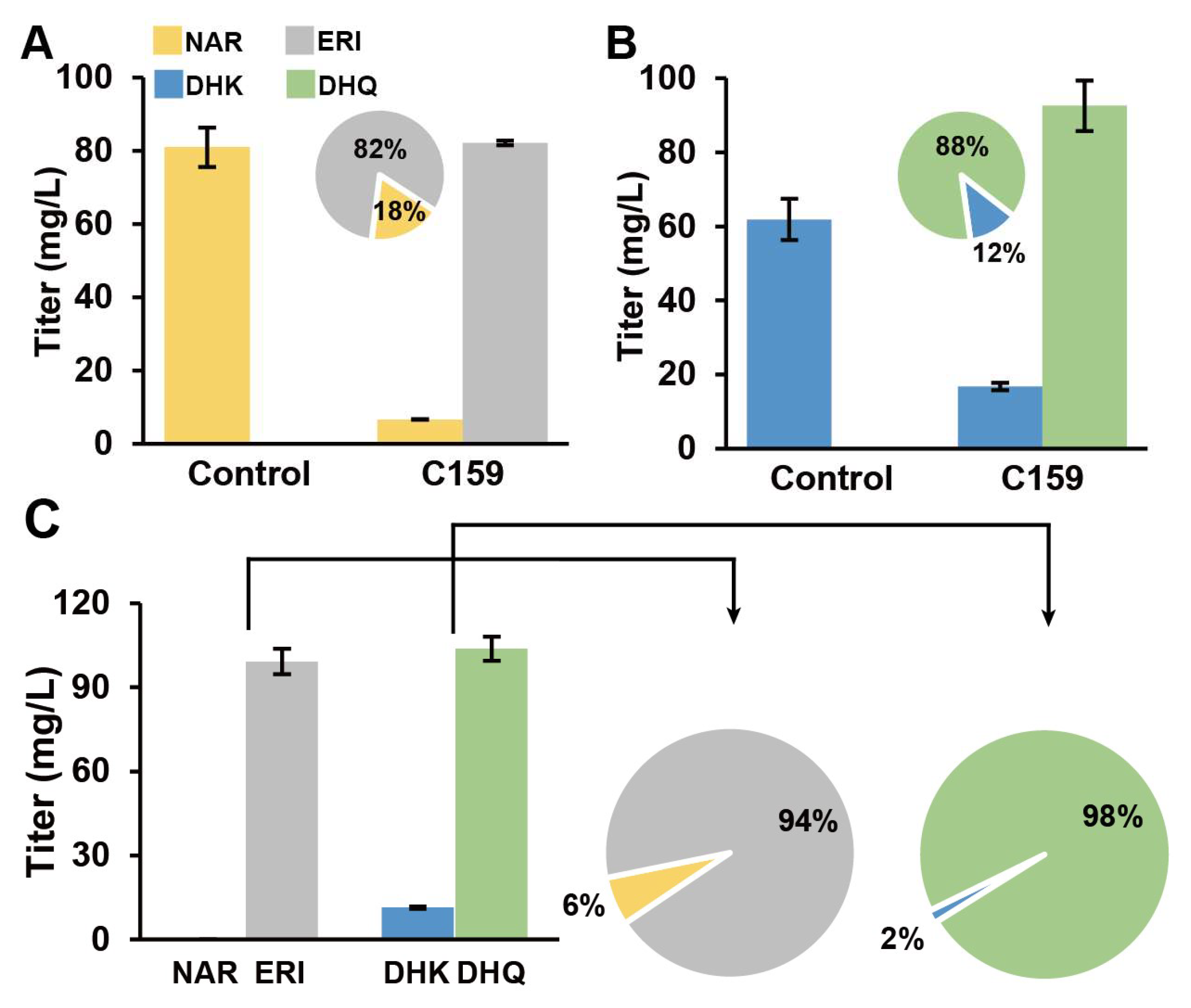
3.6. AgF3′5′H Functional Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sun, J.; Sun, W. High efficient production of plant flavonoids by microbial cell factories: Challenges and opportunities. Metab. Eng. 2022, 70, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Leonard, E.; Yan, Y. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol. Pharm. 2008, 5, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Zhao, G. Metabolic Engineering of Saccharomyces cerevisiae for De Novo Production of Kaempferol. J. Agric. Food Chem. 2019, 67, 5596–5606. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Marsafari, M. Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 2019, 8, 2514–2523. [Google Scholar] [CrossRef]
- Li, G.; Li, H. Enhanced Biosynthesis of Dihydromyricetin in Saccharomyces cerevisiae by Coexpression of Multiple Hydroxylases. J. Agric. Food Chem. 2020, 68, 14221–14229. [Google Scholar] [CrossRef]
- Wang, H.; Guo, H. Toward the Heterologous Biosynthesis of Plant Natural Products: Gene Discovery and Characterization. ACS Synth. Biol. 2021, 10, 2784–2795. [Google Scholar] [CrossRef]
- Gao, S.; Zhou, H. Promoter-Library-Based Pathway Optimization for Efficient (2S)-Naringenin Production from p-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 6884–6891. [Google Scholar] [CrossRef]
- Wang, X.; Chen, J. Engineering Escherichia coli for production of geraniol by systematic synthetic biology approaches and laboratory-evolved fusion tags. Metab. Eng. 2021, 66, 60–67. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef]
- Lv, Y.; Gao, S. Spatial organization of silybin biosynthesis in milk thistle [Silybum marianum (L.) Gaertn]. Plant J. 2017, 92, 995–1004. [Google Scholar] [CrossRef]
- Xing, M.; Cao, Y. Elucidation of myricetin biosynthesis in Morella rubra of the Myricaceae. Plant J. 2021, 108, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cao, Y. Cloning and Expression Analysis of Flavonoid 3′, 5′-Hydroxylase Gene from Brunfelsia acuminata. Genes 2021, 12, 1086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, Y. Recent advances in research on vine tea, a potential and functional herbal tea with dihydromyricetin and myricetin as major bioactive compounds. J. Pharm. Anal. 2021, 11, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Luo, F. New Method for Extracting and Purifying Dihydromyricetin from Ampelopsis grossedentata. ACS Omega 2020, 5, 13955–13962. [Google Scholar] [CrossRef]
- Hou, X.L.; Tong, Q. Suppression of Inflammatory Responses by Dihydromyricetin, a Flavonoid from Ampelopsis grossedentata, via Inhibiting the Activation of NF-kappaB and MAPK Signaling Pathways. J. Nat. Prod. 2015, 78, 1689–1696. [Google Scholar] [CrossRef]
- Liang, H.; He, K. Mechanism and antibacterial activity of vine tea extract and dihydromyricetin against Staphylococcus aureus. Sci. Rep. 2020, 10, 21416. [Google Scholar] [CrossRef]
- Wu, J.; Miyasaka, K. The Anti-Adiposity Mechanisms of Ampelopsin and Vine Tea Extract in High Fat Diet and Alcohol-Induced Fatty Liver Mouse Models. Molecules 2022, 27, 607. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y. Antioxidant activity of vine tea (Ampelopsis grossedentata) extract on lipid and protein oxidation in cooked mixed pork patties during refrigerated storage. Food Sci. Nutr. 2019, 7, 1735–1745. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Liang, X. Ampelopsin-induced autophagy protects breast cancer cells from apoptosis through Akt-mTOR pathway via endoplasmic reticulum stress. Cancer Sci. 2014, 105, 1279–1287. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, X. Dihydromyricetin improves glucose and lipid metabolism and exerts anti-inflammatory effects in nonalcoholic fatty liver disease: A randomized controlled trial. Pharmacol. Res. 2015, 99, 74–81. [Google Scholar] [CrossRef]
- Li, X.; Cao, M. Annotation of genes involved in high level of dihydromyricetin production in vine tea (Ampelopsis grossedentata) by transcriptome analysis. BMC Plant Biol. 2020, 20, 131. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.W.; Zhang, N. Exploring the genes involved in biosynthesis of dihydroquercetin and dihydromyricetin in Ampelopsis grossedentata. Sci. Rep. 2021, 11, 15596. [Google Scholar] [CrossRef]
- Park, S.; Lee, H. Functional Characterization of BrF3’H, Which Determines the Typical Flavonoid Profile of Purple Chinese Cabbage. Front. Plant Sci. 2021, 12, 793589. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, J. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018, 9, 448. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; He, X. Antioxidant Properties of a Traditional Vine Tea, Ampelopsis grossedentata. Antioxidants 2019, 8, 295. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.J.; Song, H.Y. Extracts of Vine Tea Improve Diet-Induced Non-Alcoholic Steatohepatitis Through AMPK-LXRalpha Signaling. Front. Pharmacol. 2021, 12, 711763. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, L. Cloning and functional analysis of two flavanone-3-hydroxylase genes from Reaumuria trigyna. Acta Physiol. Plant. 2014, 36, 1221–1229. [Google Scholar] [CrossRef]
- Kristine, M.; Olsen, A.H. Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol. 2010, 10, 21. [Google Scholar]
- Eichenberger, M.; Hansson, A. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, 13. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J. Cell factory engineering for improved production of natural products. Nat. Prod. Rep. 2019, 36, 1233–1236. [Google Scholar] [CrossRef]
- Jones, J.A.; Vernacchio, V.R. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab. Eng. 2016, 35, 55–63. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, D. Quercetin-derivatives paint the yellow petals of American lotus (Nelumbo lutea) and enzymatic basis for their accumulation. Hortic. Plant J. 2022, in press. [CrossRef]
- Tong, Y.; Lv, Y. Improving (2S)-naringenin production by exploring native precursor pathways and screening higher-active chalcone synthases from plants rich in flavonoids. Enzyme Microb. Technol. 2022, 156, 109991. [Google Scholar] [CrossRef]



| Gene Name | Species | Source |
|---|---|---|
| CsF3H * | Citrus sinensis | MH208416 |
| SmF3′H | Silybum marianum | [10] |
| SlF3′5′H * | Solanum lycopersicum | [5] |
| SmCPR * | S. marianum | [10] |
| ScCPR | S. cerevisiae | CP046088.1, |
| GmCPR * | Glycine max | XM_003541568.3 |
| AtCPR * | Arabidopsis thaliana | X66017.1 |
| EbCPR * | Erigeron breviscapus | [25] |
| HtCPR * | Helianthus tuberosus | Z26250.1 |
| AgF3H | A. grossedentata | This work |
| AgF3′H | A. grossedentata | This work |
| AgF3′5′H | A. grossedentata | This work |
| AgCPR1 | A. grossedentata | This work |
| AgCPR2 | A. grossedentata | This work |
| Gene | Unigene_ID | CDS Length (bp) |
|---|---|---|
| AgPAL1 | DN94338_c1_g2_i2 | 2133 |
| AgPAL2 | DN69862_c0_g1_i1 | 2172 |
| AgPAL3 | DN94338_c1_g1_i1 | 2145 |
| AgC4H | DN104001_c1_g1_i3 | 1302 |
| Ag4CL | DN87908_c0_g5_i1 | 1497 |
| AgCHS | DN99909_c4_g1_i2 | 1182 |
| AgCHI | DN86478_c0_g1_i1 | 717 |
| AgF3H | DN93644_c0_g1_i1 | 1092 |
| AgF3′H | DN153979_c0_g1_i1 | 1530 |
| AgF3′5′H | DN100751_c3_g1_i1 | 1527 |
| AgCPR1 | DN94781_c0_g1_i1 | 2118 |
| AgCPR2 | DN96223_c0_g1_i2 | 1989 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Gao, S.; Chen, Y.; Xu, S.; Yu, S.; Zhou, J. Identification of Hydroxylation Enzymes and the Metabolic Analysis of Dihydromyricetin Synthesis in Ampelopsis grossedentata. Genes 2022, 13, 2318. https://doi.org/10.3390/genes13122318
Zhang S, Gao S, Chen Y, Xu S, Yu S, Zhou J. Identification of Hydroxylation Enzymes and the Metabolic Analysis of Dihydromyricetin Synthesis in Ampelopsis grossedentata. Genes. 2022; 13(12):2318. https://doi.org/10.3390/genes13122318
Chicago/Turabian StyleZhang, Shuai, Song Gao, Yu Chen, Sha Xu, Shiqin Yu, and Jingwen Zhou. 2022. "Identification of Hydroxylation Enzymes and the Metabolic Analysis of Dihydromyricetin Synthesis in Ampelopsis grossedentata" Genes 13, no. 12: 2318. https://doi.org/10.3390/genes13122318
APA StyleZhang, S., Gao, S., Chen, Y., Xu, S., Yu, S., & Zhou, J. (2022). Identification of Hydroxylation Enzymes and the Metabolic Analysis of Dihydromyricetin Synthesis in Ampelopsis grossedentata. Genes, 13(12), 2318. https://doi.org/10.3390/genes13122318







