Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Phenotyping
2.2. Genotyping and GWAS
2.3. Candidate Gene Discovery
3. Results
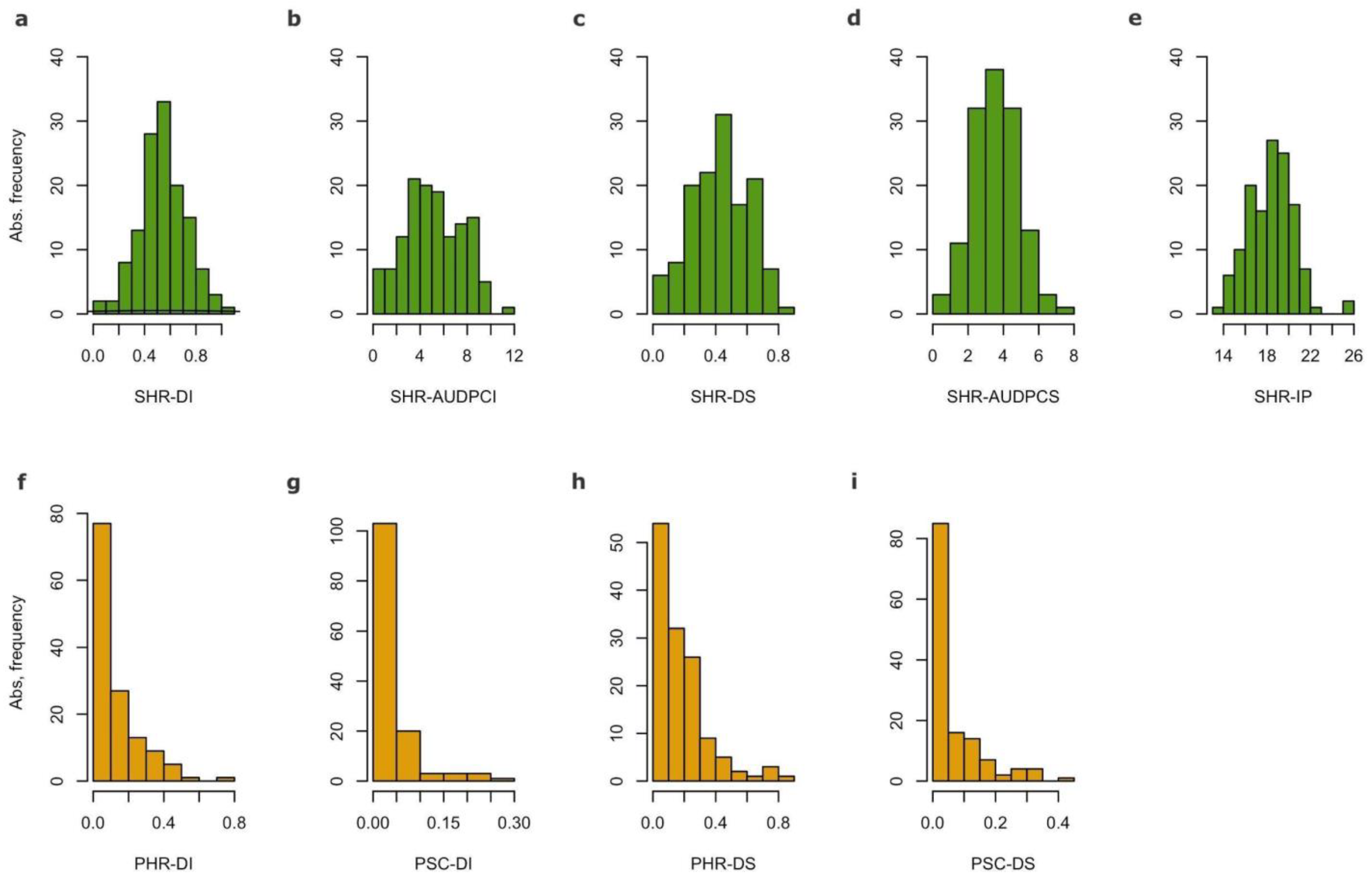
3.1. Plant Material and Phenotyping
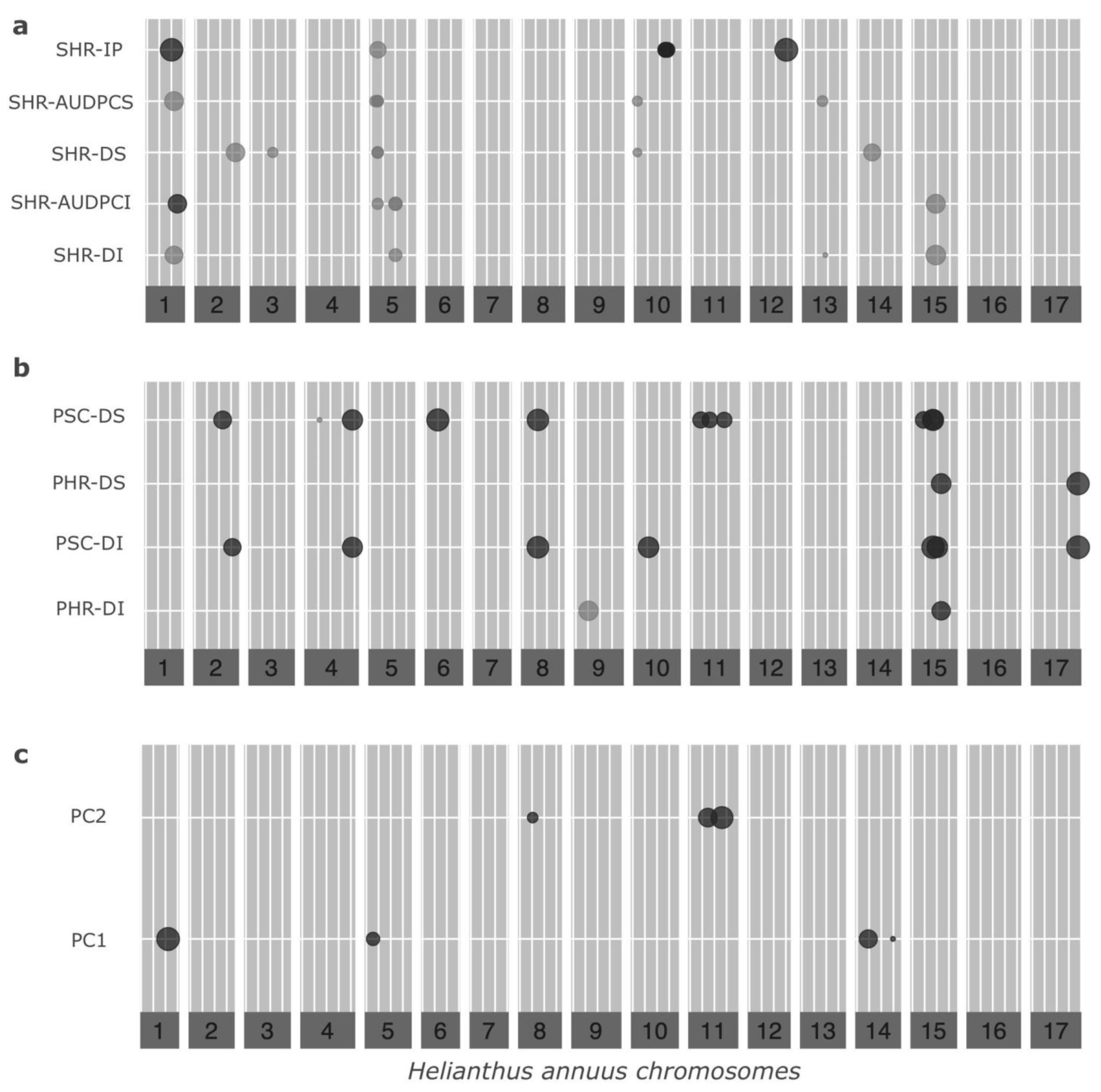
3.2. Genotyping and GWAS
3.3. Candidate Gene Discovery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA Oilseeds: World Markets and Trade. Available online: https://apps.fas.usda.gov/psdonline/circulars/oilseeds.pdf (accessed on 1 August 2022).
- Gulya, T.; Rashid, K.Y.; Masirevic, S.M. Sunflower Diseases. In Sunflower Technology and Production; Schneiter, A.A., Ed.; Agronomy Monographs; American Society of Agronomy: Madison, WI, USA; Crop Science Society of America: Madison, WI, USA; Soil Science Society of America: Madison, WI, USA, 1997; pp. 263–379. ISBN 9780891181354. [Google Scholar]
- Gulya, T.; Harveson, R.; Mathew, F.; Block, C.; Thompson, S.; Kandel, H.; Berglund, D.; Sandbakken, J.; Kleingartner, L.; Markell, S. Comprehensive disease survey of U.S. sunflower: Disease trends, research priorities and unanticipated impacts. Plant Dis. 2019, 103, 601–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masirevic, S.; Gulya, T.J. Sclerotinia and Phomopsis—Two devastating sunflower pathogens. Field Crops Res. 1992, 30, 271–300. [Google Scholar] [CrossRef]
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Derbyshire, M.C.; Newman, T.E.; Khentry, Y.; Taiwo, A.O. The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 2022, 23, 1075–1090. [Google Scholar] [CrossRef]
- Talukder, Z.I.; Underwood, W.; Misar, C.G.; Seiler, G.J.; Cai, X.; Li, X.; Qi, L. A Quantitative Genetic Study of Sclerotinia Head Rot Resistance Introgressed from the Wild Perennial Helianthus maximiliani into Cultivated Sunflower (Helianthus annuus L.). Int. J. Mol. Sci. 2022, 23, 7727. [Google Scholar] [CrossRef] [PubMed]
- Rashid, K.Y.; Block, C.C.; Gulya, T.J. Sclerotinia Head Rot and Midstalk Rot in: Compendium of Sunflower Diseases and Pests; Harveson, R.M., Markell, S.G., Block, C.C., Gulya, T.J., Eds.; The American Phytopathological Society Press: St. Paul, MN, USA, 2016; pp. 51–55. ISBN 978-0-89054-509-6. [Google Scholar]
- De Labrouhe, D.T.; Vear, F. Comparaison de méthodes d’estimation de la résistance du tournesol à Sclerotinia sclerotiorum (Lib.) de Bary. Agronomie 1984, 4, 517–525. [Google Scholar] [CrossRef]
- Fusari, C.M.; Di Rienzo, J.A.; Troglia, C.; Nishinakamasu, V.; Moreno, M.V.; Maringolo, C.; Quiroz, F.; Alvarez, D.; Escande, A.; Hopp, E.; et al. Association mapping in sunflower for Sclerotinia Head Rot resistance. BMC Plant Biol. 2012, 12, 93. [Google Scholar] [CrossRef] [Green Version]
- Filippi, C.V.; Zubrzycki, J.E.; Di Rienzo, J.A.; Quiroz, F.J.; Puebla, A.F.; Alvarez, D.; Maringolo, C.A.; Escande, A.R.; Hopp, H.E.; Heinz, R.A.; et al. Unveiling the genetic basis of Sclerotinia head rot resistance in sunflower. BMC Plant Biol. 2020, 20, 322. [Google Scholar] [CrossRef]
- Zubrzycki, J.E.; Maringolo, C.A.; Filippi, C.V.; Quiróz, F.J.; Nishinakamasu, V.; Puebla, A.F.; Di Rienzo, J.A.; Escande, A.; Lia, V.V.; Heinz, R.A.; et al. Main and epistatic QTL analyses for Sclerotinia Head Rot resistance in sunflower. PLoS ONE 2017, 12, e0189859. [Google Scholar] [CrossRef] [Green Version]
- Talukder, Z.I.; Hulke, B.S.; Qi, L.; Scheffler, B.E.; Pegadaraju, V.; McPhee, K.; Gulya, T.J. Candidate gene association mapping of Sclerotinia stalk rot resistance in sunflower (Helianthus annuus L.) uncovers the importance of COI1 homologs. Theor. Appl. Genet. 2014, 127, 193–209. [Google Scholar] [CrossRef]
- Rönicke, S.; Hahn, V.; Vogler, A.; Friedt, W. Quantitative Trait Loci Analysis of Resistance to Sclerotinia sclerotiorum in Sunflower. Phytopathology 2005, 95, 834–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muntañola-Cvetković, M.; Mihaljčević, M.; Vukojević, J.; Petrov, M. Comparisons of Phomopsis isolates obtained from sunflower plants and debris in Yugoslavia. Trans. Br. Mycol. Soc. 1985, 85, 477–483. [Google Scholar] [CrossRef]
- Acimovic, M.; Staser, N. Phomopsis sp.—A new parasite in sunflower [Helianthus annuus]. Helia 1981, 4, 43. [Google Scholar]
- Debaeke, P.; Bedoussac, L.; Bonnet, C.; Bret-Mestries, E.; Seassau, C.; Gavaland, A.; Raffaillac, D.; Tribouillois, H.; Véricel, G.; Justes, E. Sunflower crop: Environmental-friendly and agroecological. OCL 2017, 24, D304. [Google Scholar] [CrossRef] [Green Version]
- Mathew, F.; Harveson, R.; Gulya, T.; Thompson, S.; Block, C.; Markell, S. Phomopsis stem canker of sunflower. Plant Health Instr. 2018, 18. [Google Scholar] [CrossRef]
- Besnard, G.; Griveau, Y.; Quillet, M.C.; Serieys, H.; Lambert, P.; Vares, D.; Bervillé, A. Specifying the introgressed regions from H. argophyllus in cultivated sunflower (Helianthus annuus L.) to mark Phomopsis resistance genes. Theor. Appl. Genet. 1997, 94, 131–138. [Google Scholar] [CrossRef]
- Bert, P.F.; Jouan, I.; De Labrouhe, D.T.; Serre, F.; Nicolas, P.; Vear, F. Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 1. QTL involved in resistance to Sclerotinia sclerotiorum and Diaporthe helianthi. Theor. Appl. Genet. 2002, 105, 985–993. [Google Scholar] [CrossRef]
- Deglene, L.; Alibert, G.; Lesigne, P.; de Labrouhe, D.T.; Sarrafi, A. Inheritance of resistance to stem canker (Phomopsis helianthi) in sunflower. Plant Pathol. 1999, 48, 559–563. [Google Scholar] [CrossRef]
- Huguet, N. Ocurrence of phomopsis helianthi in Argentina and Uruguay. Helia 2006, 29, 121–126. [Google Scholar] [CrossRef]
- Thompson, S.M.; Tan, Y.P.; Young, A.J.; Neate, S.M.; Aitken, E.A.B.; Shivas, R.G. Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 2011, 27, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Mathew, F.M.; Alananbeh, K.M.; Jordahl, J.G.; Meyer, S.M.; Castlebury, L.A.; Gulya, T.J.; Markell, S.G. Phomopsis Stem Canker: A Reemerging Threat to Sunflower (Helianthus annuus) in the United States. Phytopathology 2015, 105, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Mathew, F.M.; Rashid, K.Y.; Gulya, T.J.; Markell, S.G. First Report of Phomopsis Stem Canker of Sunflower (Helianthus annuus) Caused by Diaporthe gulyae in Canada. Plant Dis. 2015, 99, 160. [Google Scholar] [CrossRef] [PubMed]
- Corró-Molas, A.; Comerio, R.; Figueruelo, A.; Ghironi, E. Epiphytotic disease of sunflower stem canker in Argentina. In Proceedings of the 19th International Sunflower Conference, Edirne, Turkey, 29 May–3 June 2016; pp. 825–827. [Google Scholar]
- Zambelli, A.; Mancebo, M.F.; Bazzalo, M.E.; Reid, R.J.; Sanchez, M.C.; Kontz, B.J.; Mathew, F.M. Six Species of Diaporthe Associated with Phomopsis Stem Canker of Sunflower in Southern Pampean Region of Argentina. Plant Health Prog. 2021, 22, 136–142. [Google Scholar] [CrossRef]
- Pogoda, C.S.; Reinert, S.; Talukder, Z.I.; Attia, Z.; Collier-Zans, E.C.E.; Gulya, T.J.; Kane, N.C.; Hulke, B.S. Genetic loci underlying quantitative resistance to necrotrophic pathogens Sclerotinia and Diaporthe (Phomopsis), and correlated resistance to both pathogens. Theor. Appl. Genet. 2021, 134, 249–259. [Google Scholar] [CrossRef]
- Debaeke, P.P.; Mestries, E.; Desanlis, M.; Seassau, C. Effects of crop management on the incidence and severity of fungal diseases in sunflower. In Sunflowers: Growth and Development, Environmental Influences and Pests/diseases; Arribas, J.E., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2014; ISBN 978-1-63117-348-6. [Google Scholar]
- Colombo, D.; Corro Molas, A. Relevamiento de cancro y pudrición del capítulo de girasol causado por el complejo Diaporthe. In Proceedings of the 5° Congreso Argentino de Fitopatología and 59° Reunión APS División Caribe, Cordoba, Argentina, 22–23 September 2021. [Google Scholar]
- Talukder, Z.I.; Hulke, B.S.; Marek, L.F.; Gulya, T.J. Sources of resistance to sunflower diseases in a global collection of domesticated USDA plant introductions. Crop. Sci. 2014, 54, 694–705. [Google Scholar] [CrossRef]
- Filippi, C.V.; Aguirre, N.; Rivas, J.G.; Zubrzycki, J.; Puebla, A.; Cordes, D.; Moreno, M.V.; Fusari, C.M.; Alvarez, D.; Heinz, R.A.; et al. Population structure and genetic diversity characterization of a sunflower association mapping population using SSR and SNP markers. BMC Plant Biol. 2015, 15, 52. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.O.; Montilla-Bascon, G.; Hoekenga, O.A.; Tinker, N.A.; Poland, J.; Baseggio, M.; Sorrells, M.E.; Jannink, J.-L.; Gore, M.A.; Yeats, T.H. Multivariate Genome-Wide Association Analyses Reveal the Genetic Basis of Seed Fatty Acid Composition in Oat (Avena sativa L.). G3 Genes Genomes Genet. 2019, 9, 2963–2975. [Google Scholar] [CrossRef] [Green Version]
- Filippi, C.V.; Merino, G.A.; Montecchia, J.F.; Aguirre, N.C.; Rivarola, M.; Naamati, G.; Fass, M.I.; Álvarez, D.; Di Rienzo, J.; Heinz, R.A.; et al. Genetic Diversity, Population Structure and Linkage Disequilibrium Assessment among International Sunflower Breeding Collections. Genes 2020, 11, 283. [Google Scholar] [CrossRef] [Green Version]
- Filippi, C.V.; Zubrzycki, J.E.; Di Rienzo, J.A.; Quiroz, F.; Fusari, C.M.; Alvarez, D.; Maringolo, C.A.; Cordes, D.; Escande, A.; Hopp, H.E.; et al. Phenotyping sunflower genetic resources for sclerotinia head rot response: Assessing variability for disease resistance breeding. Plant Dis. 2017, 101, 1941–1948. [Google Scholar] [CrossRef] [Green Version]
- Montecchia, J.F.; Fass, M.I.; Cerrudo, I.; Quiroz, F.J.; Nicosia, S.; Maringolo, C.A.; Di Rienzo, J.; Troglia, C.; Hopp, H.E.; Escande, A.; et al. On-field phenotypic evaluation of sunflower populations for broad-spectrum resistance to Verticillium leaf mottle and wilt. Sci. Rep. 2021, 11, 11644. [Google Scholar] [CrossRef]
- Moschen, S.; Marino, J.; Nicosia, S.; Higgins, J.; Alseekh, S.; Astigueta, F.; Bengoa Luoni, S.; Rivarola, M.; Fernie, A.R.; Blanchet, N.; et al. Exploring gene networks in two sunflower lines with contrasting leaf senescence phenotype using a system biology approach. BMC Plant Biol. 2019, 19, 446. [Google Scholar] [CrossRef]
- Moreno, M.V. Diversidad genética en girasol cultivado: Análisis de una colección de germoplasma local para su aplicación en programas de mejoramiento. Ph.D. Thesis, Universidad de Buenos Aires, Buenos Aires, Argentina, 2012. [Google Scholar]
- Schneiter, A.A.; Miller, J.F. Description of sunflower growth stages1. Crop Sci. 1981, 21, 901. [Google Scholar] [CrossRef]
- Corro Molas, A.E.; Gareis, E.; Ghironi, E. Comportamiento sanitario frente al cancro del tallo del girasol. Ensayos Comparativos de Rendimiento de Girasol 2016/2017; Colegio de Ingenieros Agrónomos de La Pampa: Santa Rosa, Argentina, 2017; pp. 12–17. [Google Scholar]
- Di Rienzo, J.; Balzarini, M.; Gonzalez, L.; Casanoves, F.; Tablada, M.; Walter Robledo, C. Infostat: Software para análisis estadístico; Universidad Nacional de Córdoba: Cordoba, Argentina, 2017. [Google Scholar]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Package “corrplot”. Statistician 2017, 56, 316–324. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. nbclust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Money, D.; Migicovsky, Z.; Gardner, K.; Myles, S. LinkImputeR: User-guided genotype calling and imputation for non-model organisms. BMC Genom. 2017, 18, 523. [Google Scholar] [CrossRef] [Green Version]
- Van Rossum, B.J.; Kruijer, W. statgenGWAS: Genome Wide Association Studies; R Package; 2020. Available online: https://cran.r-project.org/web/packages/statgenGWAS/index.html (accessed on 2 August 2022).
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [Green Version]
- Ojwang, P.P.O.; Eldridge, T.; Corredor-Moreno, P.; Njung’e, V. Structure of genetic diversity and genome-wide association studies of bean fly (Ophiomyia spencerella) resistance in common bean. Euphytica 2021, 217, 216. [Google Scholar] [CrossRef]
- Muhammad, A.; Li, J.; Hu, W.; Yu, J.; Khan, S.U.; Khan, M.H.U.; Xie, G.; Wang, J.; Wang, L. Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci. Rep. 2021, 11, 6767. [Google Scholar] [CrossRef] [PubMed]
- Carmelo, V.A.O.; Kogelman, L.J.A.; Madsen, M.B.; Kadarmideen, H.N. WISH-R- a fast and efficient tool for construction of epistatic networks for complex traits and diseases. BMC Bioinform. 2018, 19, 277. [Google Scholar] [CrossRef]
- Kogelman, L.J.A.; Kadarmideen, H.N. Weighted Interaction SNP Hub (WISH) network method for building genetic networks for complex diseases and traits using whole genome genotype data. BMC Syst. Biol. 2014, 8 (Suppl. S2), S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Programming with ggplot2. In ggplot2; Use R! Springer International Publishing: Cham, Germany, 2016; pp. 241–253. ISBN 978-3-319-24275-0. [Google Scholar]
- Osuna-Cruz, C.M.; Paytuvi-Gallart, A.; Di Donato, A.; Sundesha, V.; Andolfo, G.; Aiese Cigliano, R.; Sanseverino, W.; Ercolano, M.R. PRGdb 3.0: A comprehensive platform for prediction and analysis of plant disease resistance genes. Nucleic Acids Res. 2018, 46, D1197–D1201. [Google Scholar] [CrossRef] [PubMed]
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexa, A.; Rahnenfuhrer, J. TopGO: Enrichment Analysis for Gene Ontology, Version 2.42.0; 2006. Available online: https://bioconductor.org/packages/release/bioc/html/topGO.html (accessed on 2 August 2022).
- Mandel, J.R.; Nambeesan, S.; Bowers, J.E.; Marek, L.F.; Ebert, D.; Rieseberg, L.H.; Knapp, S.J.; Burke, J.M. Association mapping and the genomic consequences of selection in sunflower. PLoS Genet. 2013, 9, e1003378. [Google Scholar] [CrossRef] [Green Version]
- Mandel, J.R.; Dechaine, J.M.; Marek, L.F.; Burke, J.M. Genetic diversity and population structure in cultivated sunflower and a comparison to its wild progenitor, Helianthus annuus L. Theor. Appl. Genet. 2011, 123, 693–704. [Google Scholar] [CrossRef]
- Langar, K.; Griveau, Y.; Kaan, F.; Serieys, H.; Varès, D.; Bervillé, A. Evaluation of Parameters Accounting for Phomopsis Resistance Using Natural Infection and Artificial Inoculation on Recombinant Inbred Lines From a Cross Between Susceptible and Resistant Sunflower. Springer Sci. Bus. Media LLC 2002, 108, 307–315. [Google Scholar] [CrossRef]
- Hulke, B.S.; Markell, S.G.; Kane, N.C.; Mathew, F.M. Phomopsis stem canker of sunflower in North America: Correlation with climate and solutions through breeding and management. OCL 2019, 26, 13. [Google Scholar] [CrossRef]
- Aguirre, N.; Filippi, C.; Zaina, G.; Rivas, J.; Acuña, C.; Villalba, P.; García, M.; González, S.; Rivarola, M.; Martínez, M.; et al. Optimizing ddRADseq in Non-Model Species: A Case Study in Eucalyptus dunnii Maiden. Agronomy 2019, 9, 484. [Google Scholar] [CrossRef] [Green Version]
- He, L.N.; Liu, Y.J.; Xiao, P.; Zhang, L.; Guo, Y.; Yang, T.L.; Zhao, L.J.; Drees, B.; Hamilton, J.; Deng, H.Y.; et al. Genomewide linkage scan for combined obesity phenotypes using principal component analysis. Ann. Hum. Genet. 2008, 72, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, D.I.; Dolan, C.V. A comparison of power to detect a QTL in sib-pair data using multivariate phenotypes, mean phenotypes, and factor scores. Behav. Genet. 1998, 28, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Morinaka, Y.; Wang, F.; Huang, P.; Takehara, S.; Hirai, T.; Ito, A.; Koketsu, E.; Kawamura, M.; Kotake, K.; et al. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc. Natl. Acad. Sci. USA 2019, 116, 21262–21267. [Google Scholar] [CrossRef]
- Ahsan, A.; Monir, M.; Meng, X.; Rahaman, M.; Chen, H.; Chen, M. Identification of epistasis loci underlying rice flowering time by controlling population stratification and polygenic effect. DNA Res. 2019, 26, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Kim, J.-Y.; Lim, W.-J.; Jeong, S.; Lee, H.-Y.; Cho, Y.; Moon, J.-K.; Kim, N. Genome-wide association and epistatic interactions of flowering time in soybean cultivar. PLoS ONE 2020, 15, e0228114. [Google Scholar] [CrossRef] [PubMed]
- Neupane, S.; Andersen, E.J.; Neupane, A.; Nepal, M.P. Genome-Wide Identification of NBS-Encoding Resistance Genes in Sunflower (Helianthus annuus L.). Genes 2018, 9, 384. [Google Scholar] [CrossRef] [Green Version]
- Bert, P.F.; Dechamp-Guillaume, G.; Serre, F.; Jouan, I.; de Labrouhe, D.T.; Nicolas, P.; Vear, F. Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 3. Characterisation of QTL involved in resistance to Sclerotinia sclerotiorum and Phoma macdonaldi. Theor. Appl. Genet. 2004, 109, 865–874. [Google Scholar] [CrossRef]
- Guidini, R.; Braun, N.; Korah, M.; Marek, L.F.; Mathew, F.M. Greenhouse Data Suggest That Growth Stage Impacts Phomopsis Stem Canker Severity Associated with Diaporthe gulyae on Sunflower (Helianthus annuus). Plant Heal. Prog. 2021, 22, 470–473. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Grant, J.N.; Cheng, Z.-M.M.; Stewart, C.N.; Hewezi, T. Soybean kinome: Functional classification and gene expression patterns. J. Exp. Bot. 2015, 66, 1919–1934. [Google Scholar] [CrossRef]
- Rietz, S.; Bernsdorff, F.E.M.; Cai, D. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 2012, 63, 5507–5519. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, X.; Chang, X.; Sun, M.; Zhang, Y.; Li, W.; Li, Y. Overexpression of germin-like protein GmGLP10 enhances resistance to Sclerotinia sclerotiorum in transgenic tobacco. Biochem. Biophys. Res. Commun. 2018, 497, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Beracochea, V.C.; Almasia, N.I.; Peluffo, L.; Nahirñak, V.; Hopp, E.H.; Paniego, N.; Heinz, R.A.; Vazquez-Rovere, C.; Lia, V.V. Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 1717–1733. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Grimmer, J.; Pöschl, Y.; Pecher, P.; Chinchilla, D.; Scheel, D.; Lee, J. Defense-related calcium signaling mutants uncovered via a quantitative high-throughput screen in Arabidopsis thaliana. Mol. Plant 2012, 5, 115–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 2014, 9, e973818. [Google Scholar] [CrossRef] [Green Version]
- Fass, M.I.; Rivarola, M.; Ehrenbolger, G.F.; Maringolo, C.A.; Montecchia, J.F.; Quiroz, F.; García-García, F.; Blázquez, J.D.; Hopp, H.E.; Heinz, R.A.; et al. Exploring sunflower responses to Sclerotinia head rot at early stages of infection using RNA-seq analysis. Sci. Rep. 2020, 10, 13347. [Google Scholar] [CrossRef]


| Associated SNP | Chromosome | SNP Position (bp) | Disease Resistance Genes ID 1 | Class 2 | Associated Trait |
|---|---|---|---|---|---|
| X2277 | 2 | 163473796 | LOC110926750 | RLP | SHR-DS |
| X2732 | 3 | 88494320 | LOC110929054 | CK | SHR-DS |
| X4003 | 5 | 17142118 | LOC110938599, LOC118479338, LOC110938600, LOC110942752 | KIN, KIN, KIN, KIN | SHR-AUDPCS |
| X4045 | 5 | 27222485 | LOC110938841 | KIN | SHR-DS, SHR-AUDPCS |
| X4048 | 5 | 27237353 | LOC110938841, | KIN | SHR-DS, SHR-AUDPCI, SHR-AUDPCS, PC1 |
| X4053 | 5 | 28040371 | LOC110940617, LOC110940627 | KIN, KIN | SHR-IP |
| X9267 | 10 | 123241420 | LOC110882123 | N | SHR-IP |
| X9983 | 11 | 36101150 | LOC110926271 | CK | PSC-DS |
| X11154 | 12 | 143897929 | LOC110895591 | KIN | SHR-IP |
| X13326 | 15 | 42971601 | LOC110911446, LOC110911384, LOC110911385 | KIN, KIN, KIN | PSC-DS |
| X13006 | 15 | 148107847 | LOC110904282 | KIN | PC1 |
| X13502 | 15 | 91884882 | LOC110881738, LOC110881737 | KIN, KIN | SHR-DI |
| X16659 | 17 | 187789299 | LOC110926506 | KIN | PSC-DI |
| X16661 | 17 | 187789629 | LOC110926506 | KIN | PHR-DS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippi, C.V.; Corro Molas, A.; Dominguez, M.; Colombo, D.; Heinz, N.; Troglia, C.; Maringolo, C.; Quiroz, F.; Alvarez, D.; Lia, V.; et al. Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding. Genes 2022, 13, 2357. https://doi.org/10.3390/genes13122357
Filippi CV, Corro Molas A, Dominguez M, Colombo D, Heinz N, Troglia C, Maringolo C, Quiroz F, Alvarez D, Lia V, et al. Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding. Genes. 2022; 13(12):2357. https://doi.org/10.3390/genes13122357
Chicago/Turabian StyleFilippi, Carla Valeria, Andres Corro Molas, Matias Dominguez, Denis Colombo, Nicolas Heinz, Carolina Troglia, Carla Maringolo, Facundo Quiroz, Daniel Alvarez, Veronica Lia, and et al. 2022. "Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding" Genes 13, no. 12: 2357. https://doi.org/10.3390/genes13122357
APA StyleFilippi, C. V., Corro Molas, A., Dominguez, M., Colombo, D., Heinz, N., Troglia, C., Maringolo, C., Quiroz, F., Alvarez, D., Lia, V., & Paniego, N. (2022). Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding. Genes, 13(12), 2357. https://doi.org/10.3390/genes13122357








