Association of Inherited Copy Number Variation in ADAM3A and ADAM5 Pseudogenes with Oropharynx Cancer Risk and Outcome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Step 1: Screening of CNVs, Candidate DNA Region Choice, and CNVs Selection
2.3. Step 2: Validation of CNVs in OPSCC Risk
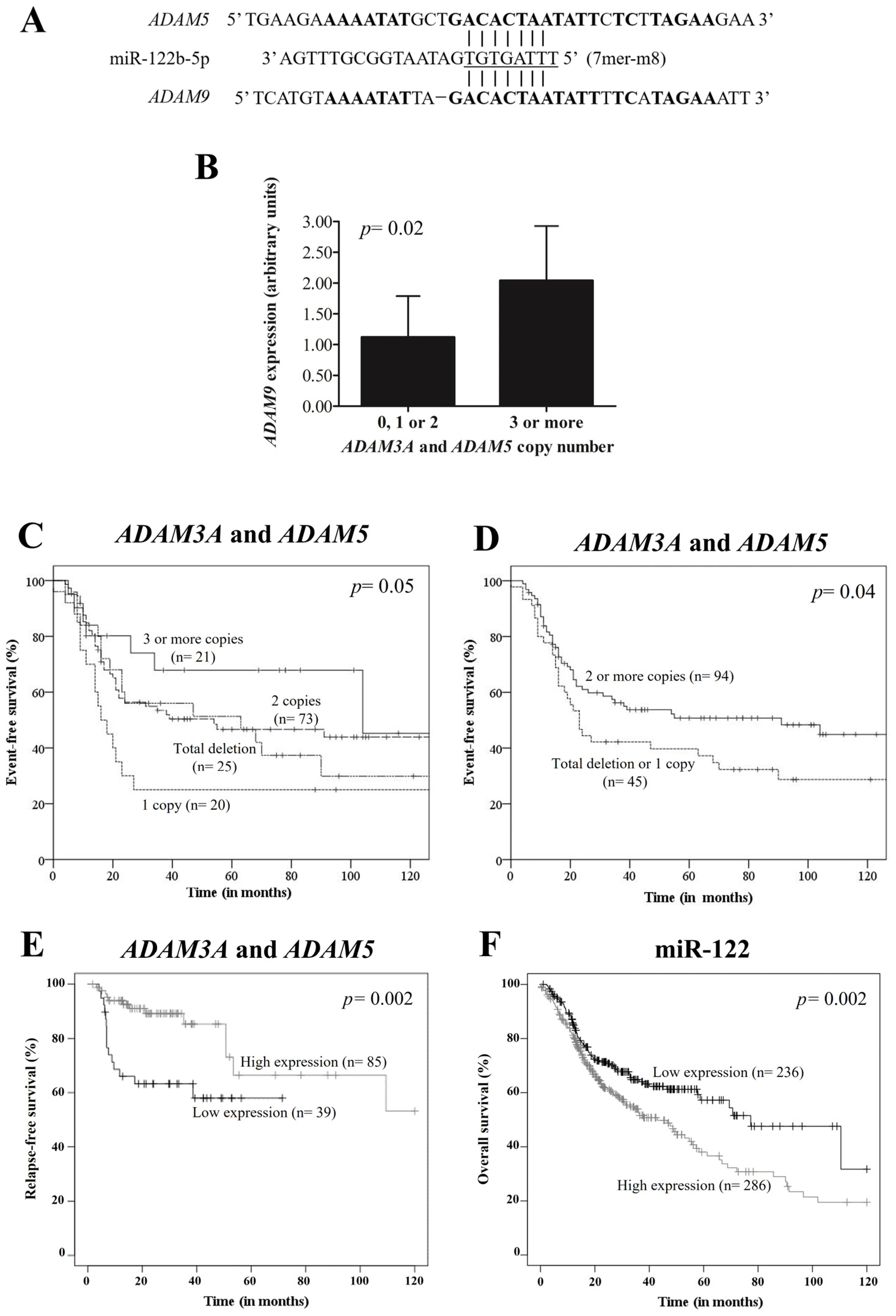
2.4. Potential Pseudogene-miRNA-mRNA Interaction
2.5. ADAM9 Expression by qPCR
2.6. Statistical Analysis
3. Results
3.1. Study Population
3.2. Step 1: Analysis of Screening and Selection of CNVs
3.3. Step 2: Analysis of Validation of CNVs
3.4. Association between Clinical and Tumor Aspects with CNVs
3.5. Potential Pseudogene-miRNA-mRNA Interaction
3.6. ADAM9 Expression by qPCR
3.7. Survival Analysis of OPSCC Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kawakita, D.; Matsuo, K. Alcohol and head and neck cancer. Cancer Metastasis Rev. 2017, 36, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Dayyani, F.; Etzel, C.J.; Liu, M.; Lippman, S.M.; Tsao, A.S. Meta-analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC). Head Neck Oncol. 2010, 2, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belcher, R.; Hayes, K.; Fedewa, S.; Chen, A.Y. Current treatment of head and neck squamous cell cancer. J. Surg. Oncol. 2014, 110, 551–574. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Sturgis, E.M.; Zhang, H.; Lu, Z.; Tao, Y.; Wei, Q.; Li, G. Genetic variants in microRNA-binding sites of DNA repair genes as predictors of recurrence in patients with squamous cell carcinoma of the oropharynx. Int. J. Cancer 2017, 141, 1355–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carron, J.; Costa, A.P.D.; Rinck-Junior, J.A.; Mariano, F.V.; Carvalho, B.S.; Lima, C.S.P.; Lourenço, G.J. Role of a genetic variation in the microRNA-4421 binding site of ERP29 regarding risk of oropharynx cancer and prognosis. Sci. Rep. 2020, 10, 17039. [Google Scholar] [CrossRef]
- Siu, L.L.; Waldron, J.N.; Chen, B.E.; Winquist, E.; Wright, J.R.; Nabid, A.; Hay, J.H.; Ringash, J.; Liu, G.; Johnson, A.; et al. Effect of Standard Radiotherapy With Cisplatin vs Accelerated Radiotherapy With Panitumumab in Locoregionally Advanced Squamous Cell Head and Neck Carcinoma: A Randomized Clinical Trial. JAMA Oncol. 2016, 3, 220–226. [Google Scholar] [CrossRef]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [Green Version]
- Krepischi, A.C.; Pearson, P.L.; Rosenberg, C. Germline copy number variations and cancer predisposition. Future Oncol. 2012, 8, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Rhie, A.; Park, W.S.; Choi, M.K.; Kim, J.R.N.; Ryu, J.; Ryu, C.H.; Kim, J.-I.; Jung, Y.-S. Genomic Copy Number Variations Characterize the Prognosis of Both P16-Positive and P16-Negative Oropharyngeal Squamous Cell Carcinoma After Curative Resection. Medicine 2015, 94, e2187. [Google Scholar] [CrossRef]
- Szyfter, K.; Wierzbicka, M.; Hunt, J.L.; Rinaldo, A.; Rodrigo, J.P.; Takes, R.P.; Ferlito, A. Frequent chromosomal aberrations and candidate genes in head and neck squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2016, 273, 537–545. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zagradišnik, B.; Krgović, D.; Herodež, Š.S.; Zagorac, A.; Ćižmarević, B.; Vokač, N.K. Identification of genomic copy number variations associated with specific clinical features of head and neck cancer. Mol. Cytogenet. 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Kempen, P.M.; Noorlag, R.; Braunius, W.W.; Moelans, C.B.; Rifi, W.; Savola, S.; Koole, R.; Grolman, W.; van Es, R.J.J.; Willems, S.M. Clinical relevance of copy number profiling in oral and oropharyngeal squamous cell carcinoma. Cancer Med. 2015, 4, 1525–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, C.H.; Guthrie, V.B.; Masica, D.L.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.M.; et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Zuo, Z.; Keck, M.K.; Khattri, A.; Pedamallu, C.S.; Stricker, T.; Brown, C.; Pugh, T.J.; Stojanov, P.; Cho, J.; et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin. Cancer Res. 2015, 21, 632–641. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Reynolds, S.; Wyczalkowski, M.A.; Oak, N.; et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018, 173, 355–370.e14. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, S.H.; Sun, W.; Zhang, K.; Liu, Y.; Nguyen, Q.; Gerasimova, A.; Nery, C.; Cheng, L.; Castonguay, C.; Hiller, E.; et al. Development and Validation of a 34-Gene Inherited Cancer Predisposition Panel Using Next-Generation Sequencing. Biomed. Res. Int. 2020, 2020, 3289023. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Liu, B.; Qiu, F.; Huang, B.; Li, Y.; Huang, D.; Yang, R.; Yang, X.; Deng, J.; Jiang, Q.; et al. The effect of functional MAPKAPK2 copy number variation CNV-30450 on elevating nasopharyngeal carcinoma risk is modulated by EBV infection. Carcinogenesis 2014, 35, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Low, J.S.; Chin, Y.M.; Mushiroda, T.; Kubo, M.; Govindasamy, G.K.; Pua, K.C.; Yap, Y.Y.; Yap, L.F.; Subramaniam, S.K.; Ong, C.A.; et al. A Genome Wide Study of Copy Number Variation Associated with Nasopharyngeal Carcinoma in Malaysian Chinese Identifies CNVs at 11q14.3 and 6p21.3 as Candidate Loci. PLoS ONE 2016, 11, e0145774. [Google Scholar] [CrossRef] [Green Version]
- Tse, K.P.; Su, W.H.; Yang, M.L.; Cheng, H.; Tsang, N.; Chang, K.; Hao, S.-P.; Shugart, Y.Y.; Chang, Y.-S. A gender-specific association of CNV at 6p21.3 with NPC susceptibility. Hum. Mol. Genet. 2011, 20, 2889–2896. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and Their miRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 2020, 8, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carron, J.; Della Coletta, R.; Lourenço, G.J. Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis. Genes 2021, 12, 1254. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xing, Y.; Xu, L.; Chen, W.; Cao, W.; Zhang, C. Decreased expression of pseudogene PTENP1 promotes malignant behaviours and is associated with the poor survival of patients with HNSCC. Sci. Rep. 2017, 7, 41179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.Z. Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene 2017, 607, 47–55. [Google Scholar] [CrossRef]
- Liu, M.; Gao, X.; Liu, C.L. Increased expression of lncRNA FTH1P3 promotes oral squamous cell carcinoma cells migration and invasion by enhancing PI3K/Akt/GSK3b/Wnt/β-catenin signaling. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8306–8314. [Google Scholar] [CrossRef]
- Yuan, H.; Jiang, H.; Wang, Y.; Dong, Y. Increased expression of lncRNA FTH1P3 predicts a poor prognosis and promotes aggressive phenotypes of laryngeal squamous cell carcinoma. Biosci. Rep. 2019, 39, BSR20181644. [Google Scholar] [CrossRef] [Green Version]
- Vincent-Chong, V.K.; Anwar, A.; Karen-Ng, L.P.; Cheong, S.C.; Yang, Y.; Pradeep, P.J.; Rahman, Z.A.A.; Ismail, S.M.; Zaini, Z.M.; Prepageran, N.; et al. Genome wide analysis of chromosomal alterations in oral squamous cell carcinomas revealed over expression of MGAM and ADAM9. PLoS ONE 2013, 8, e54705. [Google Scholar] [CrossRef]
- Rocks, N.; Paulissen, G.; El Hour, M.; Quesada, F.; Crahay, C.; Gueders, M.; Foidart, J.M.; Noel, A.; Cataldo, D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie 2008, 90, 369–379. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.A.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Glessner, J.T.; Li, J.; Hakonarson, H. ParseCNV integrative copy number variation association software with quality tracking. Nucleic Acids Res. 2013, 41, e64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. miRWalk2.0, a comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Unal, I. Defining an Optimal Cut-Point Value in ROC Analysis: An Alternative Approach. Comput. Math. Methods Med. 2017, 2017, 3762651. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masood, N.; Yasmin, A.; Kayani, M.A. Genetic variations and head and neck cancer risks. Mol. Biol. Rep. 2014, 41, 2667–2670. [Google Scholar] [CrossRef]
- Mafune, A.; Hama, T.; Suda, T.; Suzuki, Y.; Ikegami, M.; Sakanashi, C.; Imai, S.; Nakashima, A.; Yokoo, T.; Wada, K.; et al. Homozygous deletions of UGT2B17 modifies effects of smoking on TP53-mutations and relapse of head and neck carcinoma. BMC Cancer 2015, 15, 205. [Google Scholar] [CrossRef] [Green Version]
- Ambatipudi, S.; Gerstung, M.; Gowda, R.; Pai, P.; Borges, A.M.; Schäffer, A.A.; Beerenwinkel, N.; Mahimkar, M.B. Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome. PLoS ONE 2011, 6, e17250. [Google Scholar] [CrossRef] [Green Version]
- Järvinen, A.K.; Autio, R.; Kilpinen, S.; Saarela, M.; Leivo, I.; Grénman, R.; Mäkitie, A.A.; Monni, O. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer 2008, 47, 500–509. [Google Scholar] [CrossRef]
- Mochizuki, S.; Okada, Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007, 98, 621–628. [Google Scholar] [CrossRef]
- Xiao-Jie, L.; Ai-Mei, G.; Li-Juan, J.; Jiang, X. Pseudogene in cancer: Real functions and promising signature. J. Med. Genet. 2015, 52, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokes, A.; Joutsa, J.; Ala-Aho, R.; Pitchers, M.; Pennington, C.J.; Martin, C.; Premachandra, D.J.; Okada, Y.; Peltonen, J.; Grénman, R.; et al. Expression profiles and clinical correlations of degradome components in the tumor microenvironment of head and neck squamous cell carcinoma. Clin. Cancer Res. 2010, 16, 2022–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.S.; Qin, X.J.; Dai, W. Fisetin suppresses malignant proliferation in human oral squamous cell carcinoma through inhibition of Met/Src signaling pathways. Am. J. Transl. Res. 2017, 9, 5678–5683. [Google Scholar] [PubMed]
- Feng, Y.; Li, Q.; Chen, J.; Yi, P.; Xu, X.; Fan, Y.; Cui, B.; Yu, Y.; Li, X.; Du, Y.; et al. Salivary protease spectrum biomarkers of oral cancer. Int. J. Oral Sci. 2019, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.J.; Lv, L.H.; Zhang, M.; Zhou, X.; Liu, G.Q.; Lu, H.J. MiR-126 inhibits cell migration and invasion by targeting ADAM9 in oral squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10324–10331. [Google Scholar] [CrossRef]
- Salazar-Ruales, C.; Arguello, J.V.; López-Cortés, A.; Cabrera-Andrade, A.; García-Cárdenas, J.M.; Guevara-Ramírez, P.; Peralta, P.; Leone, P.E.; Paz-Y-Miño, C. Salivary MicroRNAs for Early Detection of Head and Neck Squamous Cell Carcinoma: A Case-Control Study in the High Altitude Mestizo Ecuadorian Population. Biomed. Res. Int. 2018, 2018, 9792730. [Google Scholar] [CrossRef] [Green Version]
- Liao, B.; Wang, Z.; Zhu, Y.; Wang, M.; Liu, Y. Long noncoding RNA DRAIC acts as a microRNA-122 sponge to facilitate nasopharyngeal carcinoma cell proliferation, migration and invasion via regulating SATB1. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3585–3597. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, X.; Wu, Y.L. Homozygous deletion of ADAM3A revealed by genome-wide analysis of early-stage NSCLC in China showed to be correlated with poor prognosis. J. Clin. Oncol. 2011, 29 (Suppl. 15), e21177. [Google Scholar] [CrossRef]
- Lu, J.; Zhong, Y.; Chen, J.; Lin, X.; Lin, Z.; Wang, N.; Lin, S. Radiation Enhances the Epithelial- Mesenchymal Transition of A549 Cells via miR3591-5p/USP33/PPM1A. Cell Physiol. Biochem. 2018, 50, 721–733. [Google Scholar] [CrossRef]
- Langevin, S.; Kuhnell, D.; Parry, T.; Biesiada, J.; Huang, S.; Wise-Draper, T.; Casper, K.; Zhang, X.; Medvedovic, M.; Kasper, S. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells. Oncotarget 2017, 8, 82459–82474. [Google Scholar] [CrossRef]
- Bhoi, S.; Baliakas, P.; Cortese, D.; Mattsson, M.; Engvall, M.; Smedby, K.E.; Juliusson, G.; Sutton, L.-A.; Mansouri, L. UGT2B17 expression: A novel prognostic marker within IGHV-mutated chronic lymphocytic leukemia? Haematologica 2015, 101, e63–e65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrow, J.; Adamowicz-Brice, M.; Cartmill, M.; MacArthur, D.; Lowe, J.; Robson, K.; Brundler, M.-A.; Walker, D.A.; Coyle, B.; Grundy, R. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro-Oncology 2010, 13, 212–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diels, S.; Huybreghts, S.; Van Hoorenbeeck, K.; Massa, G.; Verrijken, A.; Verhulst, S.L.; Van Gaal, L.F.; Van Hul, W. Copy number variant analysis and expression profiling of the olfactory receptor-rich 11q11 region in obesity predisposition. Mol. Genet. Metab. Rep. 2020, 25, 100656. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ise, T.; Ha, D.; Fleur, A.S.; Hahn, Y.; Liu, X.-F.; Nagata, S.; Lee, B.; Bera, T.K.; Pastan, I. Evolution and expression of chimeric POTE–actin genes in the human genome. Proc. Natl. Acad. Sci. USA 2006, 103, 17885–17890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cheng, C.; Zhang, Z.; Wang, J.; Wang, Y.; Li, X.; Gao, R.; Wang, Z.; Fang, Y.; Wang, J.; et al. Blood-based dynamic genomic signature for obsessive-compulsive disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2018, 177, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Qin, S.; Pascal, L.E.; Zou, C.; Wang, W.; Tong, H.; Zhang, J.; Catalona, W.J.; Dhir, R.; Morrell, M.; et al. SIRPB1 promotes prostate cancer cell proliferation via Akt activation. Prostate 2019, 80, 352–364. [Google Scholar] [CrossRef] [PubMed]
| CNV | OPSCC n (%) | Control n (%) | OR (CI 95%) | p Value |
|---|---|---|---|---|
| UGT2B17 (4q13.2) | ||||
| 0, 1 or 2 copies | 152 (100.0) | 150 (97.4) | NA | NA |
| 3 or more copies | 0 (0.0) | 4 (2.6) | ||
| 0 or 1 copy | 2 (1.3) | 2 (1.3) | NA | 1.00 |
| 2 or more copies | 150 (98.7) | 153 (98.7) | ||
| ADAM3A and ADAM5 (8p11.22) | ||||
| 0, 1 or 2 copies | 127 (83.6) | 148 (95.5) | Reference | 0.002 |
| 3 or more copies | 25 (16.4) | 7 (4.5) | 6.49 (1.94–21.66) | |
| 0 or 1 copy | 49 (32.2) | 47 (30.3) | Reference | 0.29 |
| 2 or more copies | 103 (67.8) | 108 (69.7) | 1.37 (0.75–2.50) | |
| SIRPB1 (20p13) | ||||
| 0, 1 or 2 copies | 1 (0.7) | 0 (0.0) | NA | NA |
| 3 or more copies | 151 (99.3) | 155 (100.0) | ||
| 0 or 1 copy | 0 (0.0) | 1 (0.6) | NA | NA |
| 2 or more copies | 152 (100.0) | 154 (99.4) |
| Characteristic | n | ADAM3A and ADAM5 CNV | |||
|---|---|---|---|---|---|
| 0, 1 or 2 n (%) | 3 or More n (%) | 0 or 1 n (%) | 2 or More n (%) | ||
| Tumor localization | 152 | ||||
| Base of tongue | 72 | 59 (81.9) | 13 (18.1) | 21 (29.2) | 51 (70.8) |
| Others | 80 | 68 (85.0) | 12 (15.0) | 28 (35.0) | 52 (65.0) |
| p value | 0.61 | 0.44 | |||
| Histological grade | 151 * | ||||
| Well or moderately | 132 | 111 (84.1) | 21 (15.9) | 45 (34.1) | 87 (65.9) |
| Poorly or undifferentiated | 19 | 15 (78.9) | 4 (21.1) | 4 (21.0) | 15 (79.0) |
| p value | 0.52 | 0.30 | |||
| Nodal stage | 152 | ||||
| 0 | 51 | 42 (82.4) | 9 (17.6) | 22 (43.1) | 29 (56.9) |
| 1 or more | 101 | 85 (84.2) | 16 (15.8) | 27 (26.7) | 74 (73.3) |
| p value | 0.81 | 0.04 ** | |||
| Tumor stage | 152 | ||||
| I-III | 49 | 43 (87.8) | 6 (12.2) | 20 (40.8) | 29 (28.2) |
| IV | 103 | 84 (81.6) | 19 (18.4) | 29 (59.2) | 74 (71.8) |
| p value | 0.48 | 0.11 | |||
| Variables | Event-Free Survival | Overall Survival | ||||
|---|---|---|---|---|---|---|
| n Event/ n Total | p Value | HR (95% CI) | n Event/ n Total | p Value | HR (95% CI) | |
| Age (years) | ||||||
| ≤57 | 48/78 | 0.40 | 1.21 (0.76–1.92) | 58/78 | 0.27 | 1.24 (0.84–1.82) |
| >57 | 29/61 | Reference | 48/61 | Reference | ||
| Gender | ||||||
| Female | 0/5 | 0.16 | Reference | 2/5 | 0.10 | Reference |
| Male | 77/134 | 22.08 (NA) | 104/134 | 3.23 (0.79–13.11) | ||
| Histological grade | ||||||
| Poor | 11/19 | 0.54 | 1.22 (0.64–2.31) | 14/19 | 0.81 | 1.07 (0.60–1.88) |
| Moderate | 66/120 | Reference | 92/120 | Reference | ||
| TNM stage | ||||||
| I–III | 18/44 | 0.003 * | Reference | 29/44 | 0.001 | Reference |
| IV | 59/95 | 2.24 (1.31–3.82) | 77/95 | 2.12 (1.37–3.26) | ||
| ADAM3A and ADAM5 | ||||||
| 0, 1 or 2 copies | 70/118 | 0.14 | 1.78 (0.82–3.88) | 90/118 | 0.77 | 1.08 (0.63–1.84) |
| 3 or more copies | 7/21 | Reference | 16/21 | Reference | ||
| 0 or 1 copy | 32/45 | 0.04 ** | 1.58 (1.01–2.48) | 32/45 | 0.69 | 1.08 (0.71–1.64) |
| 2 or more copies | 45/94 | Reference | 74/94 | Reference | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carron, J.; Torricelli, C.; Silva, J.K.; Liu, Y.; Pellegrino, R.; Lima, C.S.P.; Lourenço, G.J. Association of Inherited Copy Number Variation in ADAM3A and ADAM5 Pseudogenes with Oropharynx Cancer Risk and Outcome. Genes 2022, 13, 2408. https://doi.org/10.3390/genes13122408
Carron J, Torricelli C, Silva JK, Liu Y, Pellegrino R, Lima CSP, Lourenço GJ. Association of Inherited Copy Number Variation in ADAM3A and ADAM5 Pseudogenes with Oropharynx Cancer Risk and Outcome. Genes. 2022; 13(12):2408. https://doi.org/10.3390/genes13122408
Chicago/Turabian StyleCarron, Juliana, Caroline Torricelli, Janet Keller Silva, Yichuan Liu, Renata Pellegrino, Carmen Silvia Passos Lima, and Gustavo Jacob Lourenço. 2022. "Association of Inherited Copy Number Variation in ADAM3A and ADAM5 Pseudogenes with Oropharynx Cancer Risk and Outcome" Genes 13, no. 12: 2408. https://doi.org/10.3390/genes13122408





