Increase in Ribosomal Fidelity Benefits Salmonella upon Bile Salt Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Growth Conditions
2.2. Determination of Mistranslation Rates
2.3. Bacterial Dynamic Growth Curve
2.4. Liquid Media Competition Assays
2.5. Measurement of Intracellular ATP
2.6. Quantitation and Statistical Analysis
3. Results
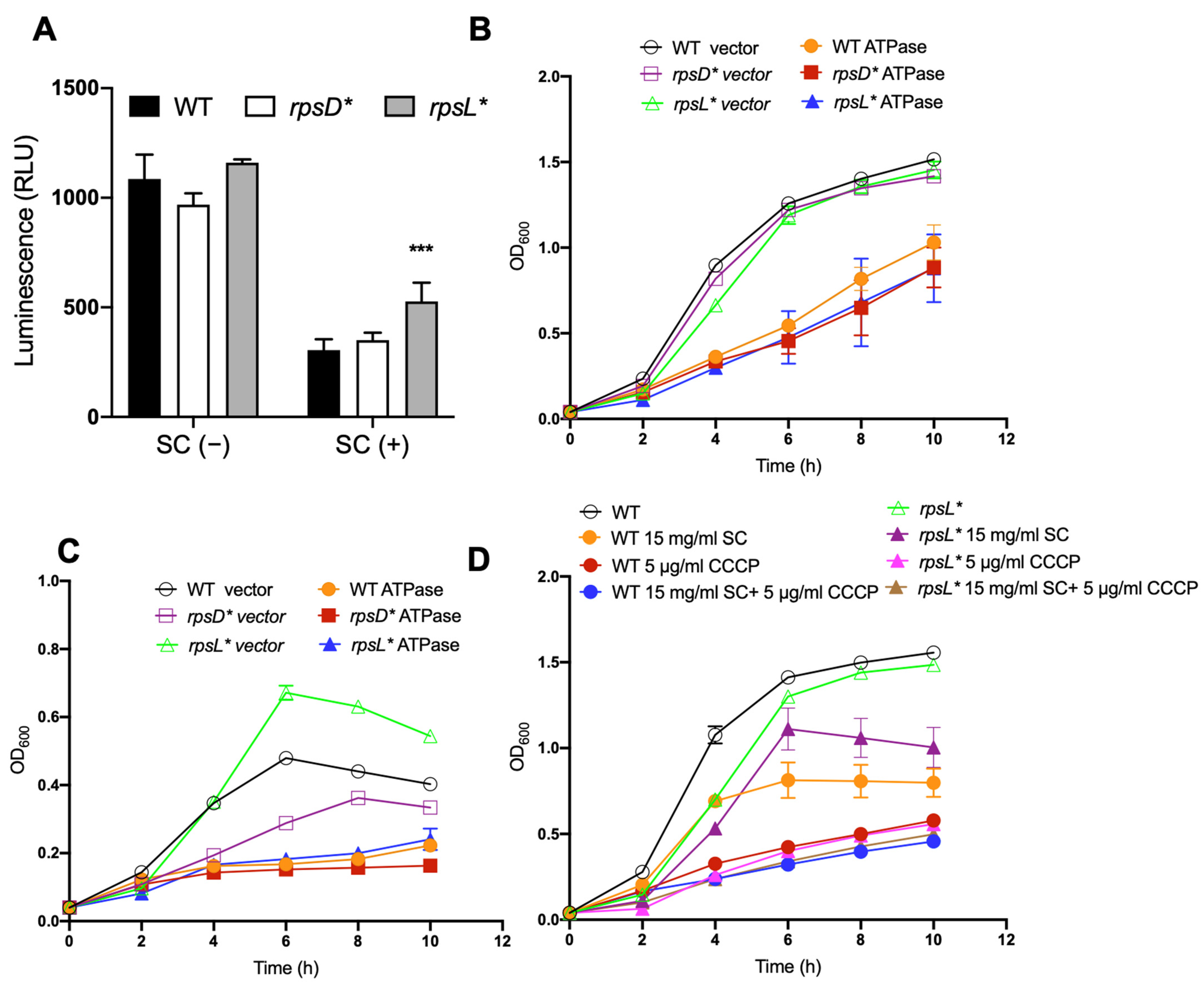
3.1. Higher Translational Fidelity Increases Fitness upon Bile Salt Exposure
3.2. Porins, Envelope Stress Response, and Flagella Are Not Major Contributors to Improved Bile Salt Resistance
3.3. Improved Fitness in High-Fidelity Strain Depend on Increased Intracellular ATP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurland, C.G. Translational Accuracy and the Fitness of Bacteria. Annu. Rev. Genet. 1992, 26, 29–50. [Google Scholar] [CrossRef]
- Kramer, E.B.; Vallabhaneni, H.; Mayer, L.M.; Farabaugh, P.J. A Comprehensive Analysis of Translational Missense Errors in the Yeast Saccharomyces Cerevisiae. RNA 2010, 16, 1797–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, J.; So, B.R.; Yadavalli, S.S.; Roy, H.; Shoji, S.; Fredrick, K.; Musier-Forsyth, K.; Ibba, M. Resampling and Editing of Mischarged TRNA Prior to Translation Elongation. Mol. Cell 2009, 33, 654–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traverse, C.C.; Ochman, H. Conserved Rates and Patterns of Transcription Errors across Bacterial Growth States and Lifestyles. Proc. Natl. Acad. Sci. USA 2016, 113, 3311–3316. [Google Scholar] [CrossRef] [Green Version]
- Gordon, A.J.E.; Satory, D.; Halliday, J.A.; Herman, C. Lost in Transcription: Transient Errors in Information Transfer. Curr. Opin. Microbiol. 2015, 24, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Gromadski, K.B.; Rodnina, M.V. Kinetic Determinants of High-Fidelity TRNA Discrimination on the Ribosome. Mol. Cell 2004, 13, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Boniecki, M.T.; Jaffe, J.D.; Imai, B.S.; Yau, P.M.; Luthey-Schulten, Z.A.; Martinis, S.A. Naturally Occurring Aminoacyl-TRNA Synthetases Editing-Domain Mutations That Cause Mistranslation in Mycoplasma Parasites. Proc. Natl. Acad. Sci. USA 2011, 108, 9378–9383. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Vargas-Rodriguez, O.; Goto, Y.; Novoa, E.M.; Ribas de Pouplana, L.; Suga, H.; Musier-Forsyth, K. Homologous Trans-Editing Factors with Broad TRNA Specificity Prevent Mistranslation Caused by Serine/Threonine Misactivation. Proc. Natl. Acad. Sci. USA 2015, 112, 6027–6032. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, M.H.; Pan, T. Temperature Dependent Mistranslation in a Hyperthermophile Adapts Proteins to Lower Temperatures. Nucleic Acids Res. 2016, 44, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Gromadski, K.B.; Rodnina, M.V. Streptomycin Interferes with Conformational Coupling between Codon Recognition and GTPase Activation on the Ribosome. Nat. Struct. Mol. Biol. 2004, 11, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Bullwinkle, T.J.; Reynolds, N.M.; Raina, M.; Moghal, A.; Matsa, E.; Rajkovic, A.; Kayadibi, H.; Fazlollahi, F.; Ryan, C.; Howitz, N.; et al. Oxidation of Cellular Amino Acid Pools Leads to Cytotoxic Mistranslation of the Genetic Code. eLife 2014, 3, e02501. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Wierzbowski, J.; Cottarel, G.; Collins, J.J. Mistranslation of Membrane Proteins and Two-Component System Activation Trigger Antibiotic-Mediated Cell Death. Cell 2008, 135, 679–690. [Google Scholar] [CrossRef] [Green Version]
- Lant, J.T.; Kiri, R.; Duennwald, M.L.; O’Donoghue, P. Formation and Persistence of Polyglutamine Aggregates in Mistranslating Cells. Nucleic Acids Res. 2021, 49, 11883–11899. [Google Scholar] [CrossRef] [PubMed]
- Mikkola, R.; Kurland, C.G. Selection of Laboratory Wild-Type Phenotype from Natural Isolates of Escherichia Coli in Chemostats. Mol. Biol. Evol. 1992, 9, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, A.R.; Simões, J.; Lee, W.; Rung, J.; Weil, T.; Gut, I.G.; Gut, M.; Bayés, M.; Rizzetto, L.; Cavalieri, D.; et al. Reversion of a Fungal Genetic Code Alteration Links Proteome Instability with Genomic and Phenotypic Diversification. Proc. Natl. Acad. Sci. USA 2013, 110, 11079–11084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, T. Adaptive Translation as a Mechanism of Stress Response and Adaptation. Annu. Rev. Genet. 2013, 47, 121–137. [Google Scholar] [CrossRef] [Green Version]
- Ribas de Pouplana, L.; Santos, M.A.S.; Zhu, J.-H.; Farabaugh, P.J.; Javid, B. Protein Mistranslation: Friend or Foe? Trends Biochem. Sci. 2014, 39, 355–362. [Google Scholar] [CrossRef]
- Fan, Y.; Wu, J.; Ung, M.H.; De Lay, N.; Cheng, C.; Ling, J. Protein Mistranslation Protects Bacteria against Oxidative Stress. Nucleic Acids Res. 2015, 43, 1740–1748. [Google Scholar] [CrossRef]
- Mohler, K.; Ibba, M. Translational Fidelity and Mistranslation in the Cellular Response to Stress. Nat. Microbiol. 2017, 2, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Thompson, L.; Lyu, Z.; Cameron, T.A.; De Lay, N.R.; Krachler, A.M.; Ling, J. Optimal Translational Fidelity Is Critical for Salmonella Virulence and Host Interactions. Nucleic Acids Res. 2019, 47, 5356–5367. [Google Scholar] [CrossRef] [PubMed]
- Andrews-Polymenis, H.L.; Bäumler, A.J.; McCormick, B.A.; Fang, F.C. Taming the Elephant: Salmonella Biology, Pathogenesis, and Prevention. Infect. Immun. 2010, 78, 2356–2369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and Acute Infection of the Gall Bladder by Salmonella Typhi: Understanding the Carrier State. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontes, M.H.; Groisman, E.A. Protein Synthesis Controls Phosphate Homeostasis. Genes Dev. 2018, 32, 79–92. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Evans, C.R.; Barber, K.W.; Banerjee, K.; Weiss, K.J.; Margolin, W.; Igoshin, O.A.; Rinehart, J.; Ling, J. Heterogeneity of Stop Codon Readthrough in Single Bacterial Cells and Implications for Population Fitness. Mol. Cell 2017, 67, 826–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatfield, S.N.; Dorman, C.J.; Hayward, C.; Dougan, G. Role of OmpR-Dependent Genes in Salmonella Typhimurium Virulence: Mutants Deficient in Both OmpC and OmpF Are Attenuated in Vivo. Infect. Immun. 1991, 59, 449–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slauch, J.M.; Silhavy, T.J. Genetic Analysis of the Switch That Controls Porin Gene Expression in Escherichia Coli K-12. J. Mol. Biol. 1989, 210, 281–292. [Google Scholar] [CrossRef]
- Slauch, J.M.; Garrett, S.; Jackson, D.E.; Silhavy, T.J. EnvZ Functions through OmpR to Control Porin Gene Expression in Escherichia Coli K-12. J. Bacteriol. 1988, 170, 439–441. [Google Scholar] [CrossRef] [Green Version]
- Heyde, M.; Portalier, R. Regulation of Major Outer Membrane Porin Proteins of Escherichia Coli K 12 by PH. Mol. Gen. Genet. 1987, 208, 511–517. [Google Scholar] [CrossRef]
- Gunn, J.S. Mechanisms of Bacterial Resistance and Response to Bile. Microbes Infect 2000, 2, 907–913. [Google Scholar] [CrossRef]
- Picken, R.N.; Beacham, I.R. Bacteriophage-Resistant Mutants of Escherichia Coli K12. Location of Receptors within the Lipopolysaccharide. J. Gen. Microbiol. 1977, 102, 305–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, S.B.; Cava, F.; Pucciarelli, M.G.; García-Del Portillo, F.; de Pedro, M.A.; Casadesús, J. Bile-Induced Peptidoglycan Remodelling in Salmonella Enterica. Environ. Microbiol. 2015, 17, 1081–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.M.; Silhavy, T.J. Envelope Stress Responses: Balancing Damage Repair and Toxicity. Nat. Rev. Microbiol. 2019, 17, 417–428. [Google Scholar] [CrossRef]
- Merritt, M.E.; Donaldson, J.R. Effect of Bile Salts on the DNA and Membrane Integrity of Enteric Bacteria. J. Med. Microbiol. 2009, 58, 1533–1541. [Google Scholar] [CrossRef] [Green Version]
- Lyu, Z.; Yang, A.; Villanueva, P.; Singh, A.; Ling, J. Heterogeneous Flagellar Expression in Single Salmonella Cells Promotes Diversity in Antibiotic Tolerance. MBio 2021, 12, e02374-21. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Cho, C.; Guo, L.-T.; Aerni, H.R.; Rinehart, J.; Söll, D. Protein Aggregation Caused by Aminoglycoside Action Is Prevented by a Hydrogen Peroxide Scavenger. Mol. Cell 2012, 48, 713–722. [Google Scholar] [CrossRef] [Green Version]
- Begley, M.; Gahan, C.G.M.; Hill, C. The Interaction between Bacteria and Bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [Green Version]
- Baneyx, F.; Mujacic, M. Recombinant Protein Folding and Misfolding in Escherichia Coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef]
- Chen, I.; Cassaro, S. Physiology, Bile salts. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same Species, Different Diseases: How and Why Typhoidal and Non-Typhoidal Salmonella Enterica Serovars Differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef] [Green Version]
- Stanaway, J.D.; Parisi, A.; Sarkar, K.; Blacker, B.F.; Reiner, R.C.; Hay, S.I.; Nixon, M.R.; Dolecek, C.; James, S.L.; Mokdad, A.H.; et al. GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators The Global Burden of Non-Typhoidal Salmonella Invasive Disease: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 1312–1324. [Google Scholar] [CrossRef] [Green Version]
- LaRock, D.L.; Chaudhary, A.; Miller, S.I. Salmonellae Interactions with Host Processes. Nat. Rev. Microbiol. 2015, 13, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Menendez, A.; Arena, E.T.; Guttman, J.A.; Thorson, L.; Vallance, B.A.; Vogl, W.; Finlay, B.B. Salmonella Infection of Gallbladder Epithelial Cells Drives Local Inflammation and Injury in a Model of Acute Typhoid Fever. J. Infect. Dis. 2009, 200, 1703–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biofilm Formation and Interaction with the Surfaces of Gallstones by Salmonella spp. | Infection and Immunity. Available online: https://journals.asm.org/doi/10.1128/IAI.70.5.2640-2649.2002 (accessed on 28 December 2021).
- Gallstones Play a Significant Role in Salmonella spp. Gallbladder Colonization and Carriage | PNAS. Available online: https://www.pnas.org/content/107/9/4353 (accessed on 28 December 2021).
- Lee, J.Y.; Kim, D.G.; Kim, B.-G.; Yang, W.S.; Hong, J.; Kang, T.; Oh, Y.S.; Kim, K.R.; Han, B.W.; Hwang, B.J.; et al. Promiscuous Methionyl-TRNA Synthetase Mediates Adaptive Mistranslation to Protect Cells against Oxidative Stress. J. Cell Sci. 2014, 127, 4234–4245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javid, B.; Sorrentino, F.; Toosky, M.; Zheng, W.; Pinkham, J.T.; Jain, N.; Pan, M.; Deighan, P.; Rubin, E.J. Mycobacterial Mistranslation Is Necessary and Sufficient for Rifampicin Phenotypic Resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 1132–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thanassi, D.G.; Cheng, L.W.; Nikaido, H. Active Efflux of Bile Salts by Escherichia Coli. J. Bacteriol. 1997, 179, 2512–2518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, E.Y.; Bertenthal, D.; Nilles, M.L.; Bertrand, K.P.; Nikaido, H. Bile Salts and Fatty Acids Induce the Expression of Escherichia Coli AcrAB Multidrug Efflux Pump through Their Interaction with Rob Regulatory Protein. Mol. Microbiol. 2003, 48, 1609–1619. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Dazer, M.; Miller, P.F. The Salmonella Typhimurium Mar Locus: Molecular and Genetic Analyses and Assessment of Its Role in Virulence. J. Bacteriol. 1997, 179, 1857–1866. [Google Scholar] [CrossRef] [Green Version]
- van Velkinburgh, J.C.; Gunn, J.S. PhoP-PhoQ-Regulated Loci Are Required for Enhanced Bile Resistance in Salmonella spp. Infect Immun. 1999, 67, 1614–1622. [Google Scholar] [CrossRef]



| Strains | Source or Reference | Genotype/Features |
|---|---|---|
| S. typhimurium 14,028 s (WT) | ATCC | N/A |
| rpsD* | Lab collection | rpsD I199N |
| rpsL* | Lab collection | rpsL K42N |
| rpsD*L* | Lab collection | rpsD I199N and rpsL K42N |
| ΔompF:kan | This study | Region 1,048,143 to 1,049,228 (Δ2–363 aa) |
| ΔompC:cat | This study | Region 2,416,999 to 2,418,129 (Δ2–378 aa) |
| ΔompR:cat | This study | Region 3,673,507 to 3,674,220 (Δ2–239 aa) |
| ΔcpxR:cat | This study | Region 4,283,333 to 4,284,025 (Δ2–232 aa) |
| ΔflhDC:cat | Lab collection | Region 2,032,540 to 2,033,471 (ΔflhD 1–117 aa ΔflhC 1–193 aa) |
| Plasmids | Source or Reference | Genotype/Features |
| pKD46 | Lab collection | ampR |
| pKD3 | Lab collection | ampR and camR |
| pKD4 | Lab collection | ampR and kanR |
| pZS-Ptet-m-y | Lab collection | ampR |
| pZS-Ptet-m-TGA-y | Lab collection | ampR |
| pZS-Ptet-lacZ | Lab collection | ampR |
| pZS-Ptet-YFP | Lab collection | ampR |
| pZS-Ptet-eCFP | Lab collection | ampR |
| pUHE-ATPase | [24] | ampR |
| pVector | [24] | ampR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, Z.; Ling, J. Increase in Ribosomal Fidelity Benefits Salmonella upon Bile Salt Exposure. Genes 2022, 13, 184. https://doi.org/10.3390/genes13020184
Lyu Z, Ling J. Increase in Ribosomal Fidelity Benefits Salmonella upon Bile Salt Exposure. Genes. 2022; 13(2):184. https://doi.org/10.3390/genes13020184
Chicago/Turabian StyleLyu, Zhihui, and Jiqiang Ling. 2022. "Increase in Ribosomal Fidelity Benefits Salmonella upon Bile Salt Exposure" Genes 13, no. 2: 184. https://doi.org/10.3390/genes13020184
APA StyleLyu, Z., & Ling, J. (2022). Increase in Ribosomal Fidelity Benefits Salmonella upon Bile Salt Exposure. Genes, 13(2), 184. https://doi.org/10.3390/genes13020184







