Identification and Functional Analysis of the Regulatory Elements in the pHSPA6 Promoter
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. Plasmids
2.3. Electroporation of H2E Cells
2.4. Selection of Cell Clones
2.5. PCR Detection
2.6. Quantitative Real-Time PCR Analysis
2.7. Fluorescence Microscopy and Flow Cytometric Analysis
2.8. Statistical Analysis
3. Results
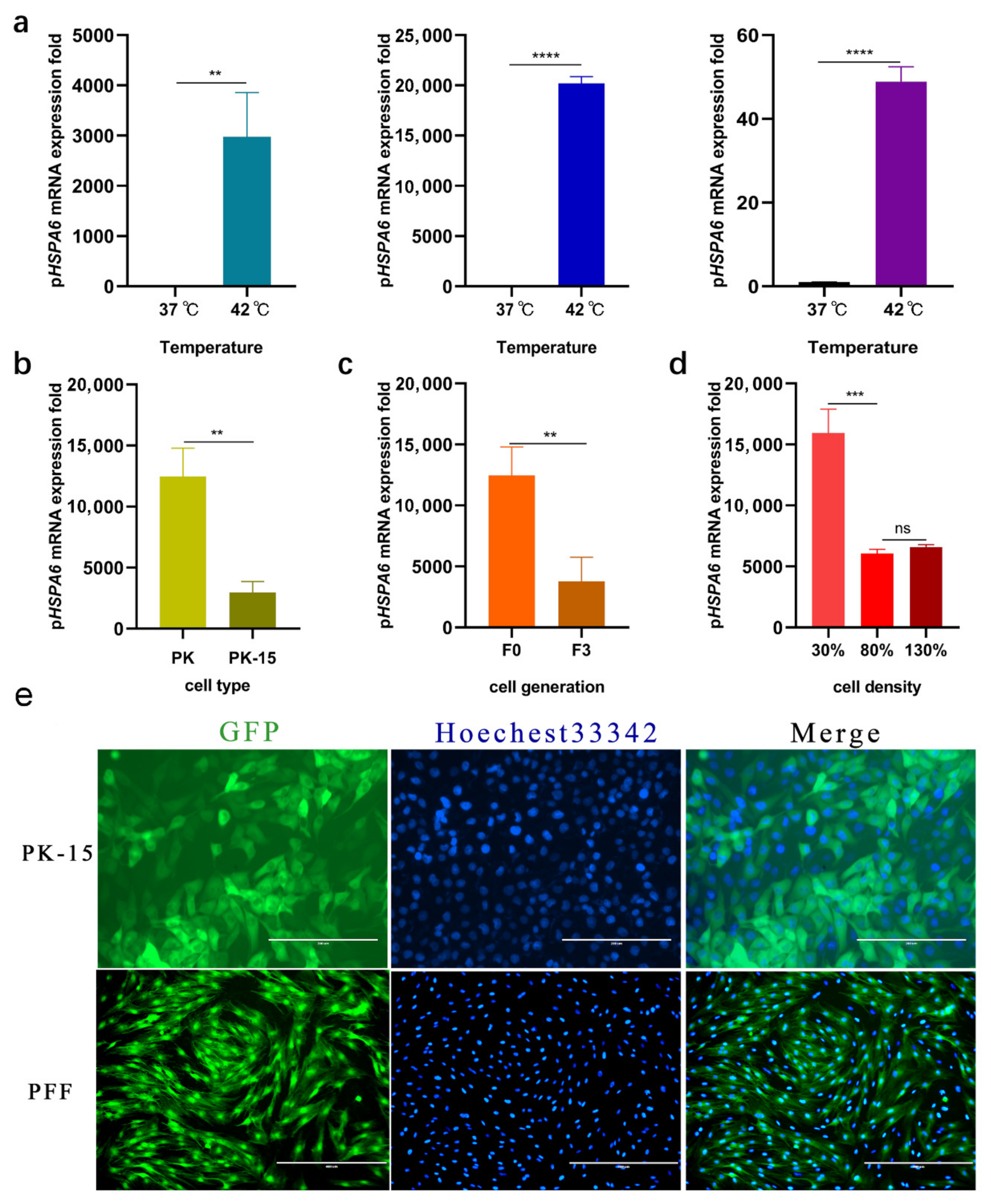
3.1. Expression of HSPA6 in Porcine Cells
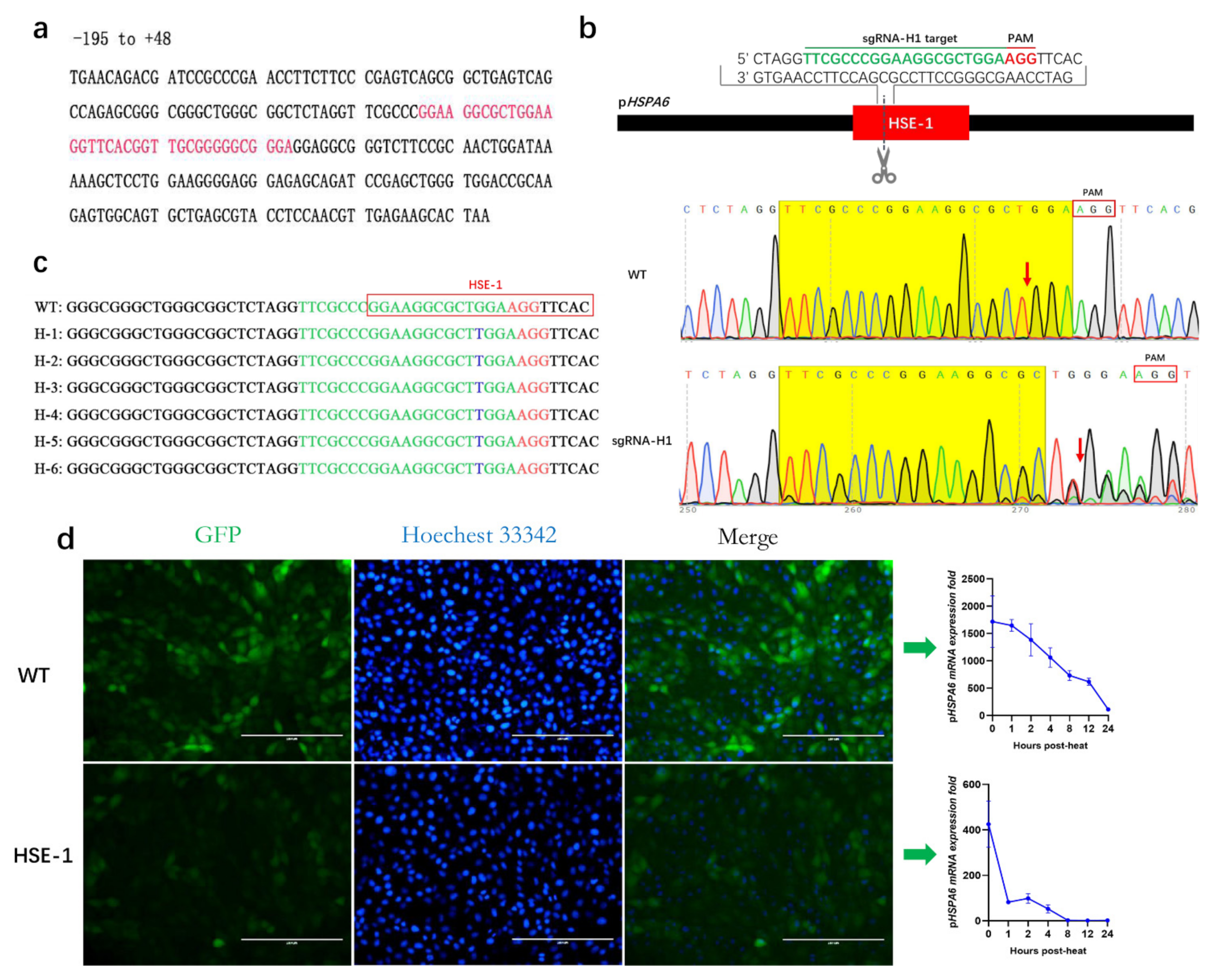
3.2. Effect of HSE-1 on the Expression of pHSPA6
3.3. Effect of HSE-2 on the Expression of pHSPA6
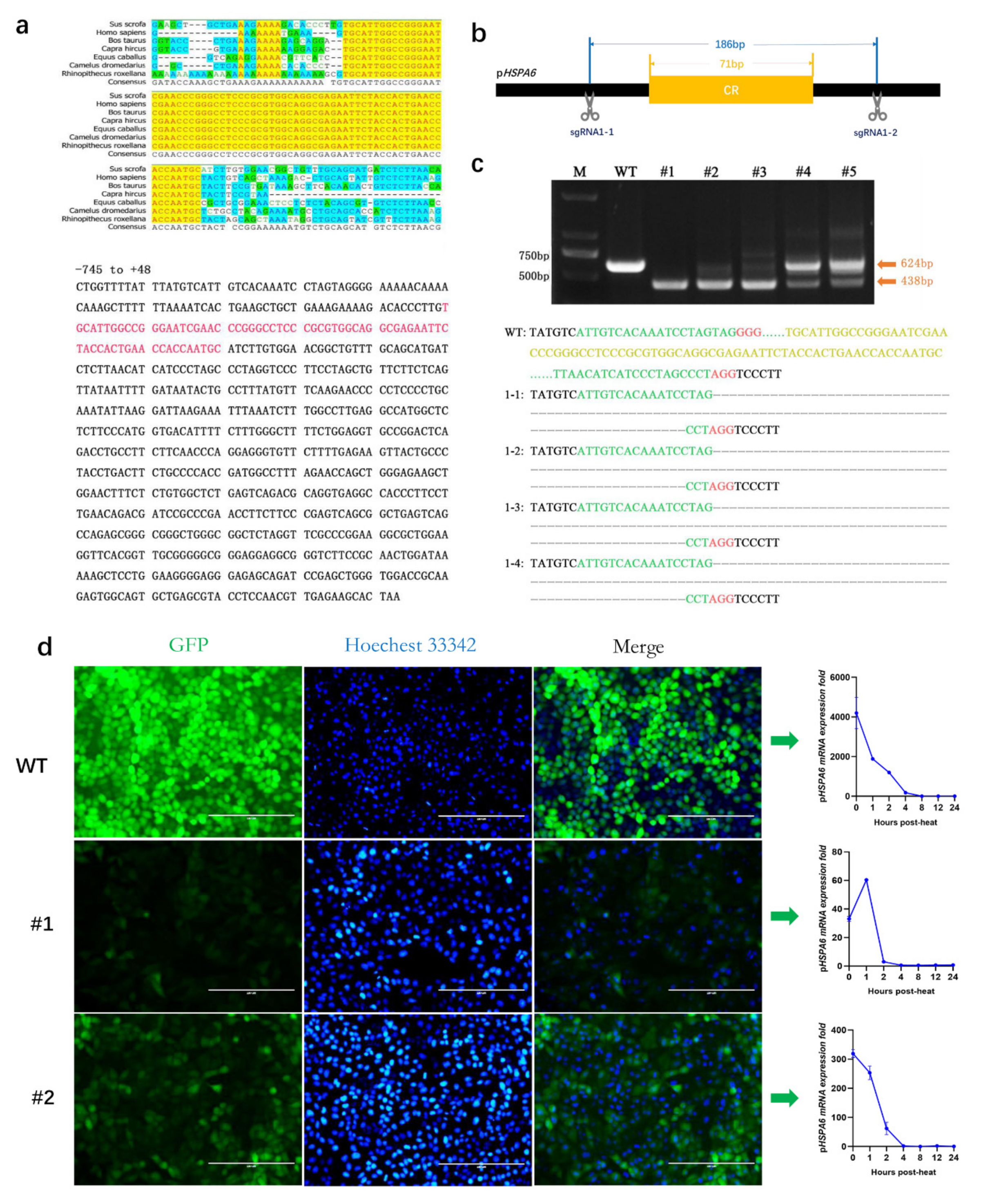
3.4. Effect of the CR on the Expression of pHSPA6
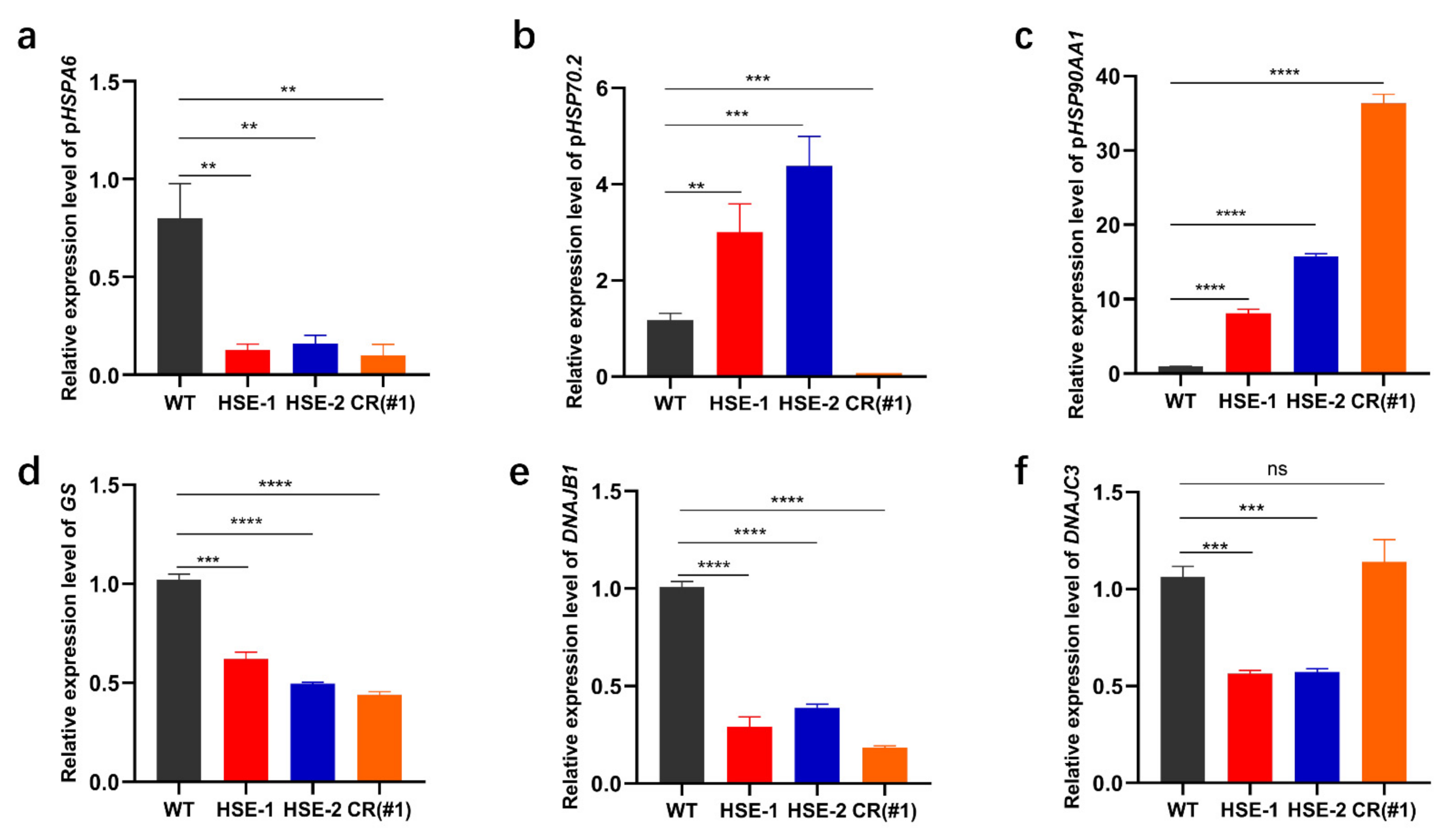
3.5. Effects of the Three Regulatory Regions on the Expression of Other Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nitika; Porter, C.M.; Truman, A.W.; Truttmann, M.C. Post-translational modifications of Hsp70 family proteins: Expanding the chaperone code. J. Biol. Chem. 2020, 295, 10689–10708. [Google Scholar] [CrossRef]
- Noonan, E.J.; Place, R.F.; Giardina, C.; Hightower, L.E. Hsp70B′ regulation and function. Cell Stress Chaperon. 2007, 12, 219–229. [Google Scholar] [CrossRef]
- Xu, Q.; Schett, G.; Li, C.; Hu, Y.; Wick, G. Mechanical stress-induced heat shock protein 70 expression in vascular smooth muscle cells is regulated by Rac and Ras small G proteins but not mitogen-activated protein kinases. Circ. Res. 2000, 86, 1122–1128. [Google Scholar] [CrossRef]
- Hyder, I.; Sejian, V.; Bhatta, R.; Gaughan, J.B. Biological role of melatonin during summer season related heat stress in livestock. Biol. Rhythm. Res. 2017, 48, 297–314. [Google Scholar] [CrossRef]
- Banerjee, D.; Upadhyay, R.C.; Chaudhary, U.B.; Kumar, R.; Singh, S.; Ashutosh, G.J.M.; Polley, S.; Mukherjee, A.; Das, T.K.; De, S. Seasonal variation in expression pattern of genes under HSP70. Cell Stress Chaperon. 2014, 19, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Mohanarao, G.J.; Mukherjee, A.; Banerjee, D.; Gohain, M.; Dass, G.; Brahma, B.; Datta, T.K.; Upadhyay, R.C.; De, S. HSP70 family genes and HSP27 expression in response to heat and cold stress in vitro in peripheral blood mononuclear cells of goat (Capra hircus). Small Rumin. Res. 2014, 116, 94–99. [Google Scholar] [CrossRef]
- Rocha, R.; Baena, M.M.; Estopa, A.D.C.; Gervásio, I.C.; Ibelli, A.M.G.; Gionbelli, T.R.S.; Gionbelli, M.P.; De Freitas, R.T.F.; Meirelles, S.L.C. Differential expression of HSF1 and HSPA6 genes and physiological responses in Angus and Simmental cattle breeds. J. Therm. Biol. 2019, 84, 92–98. [Google Scholar] [CrossRef]
- Hageman, J.; Van Waarde-Verhagen, M.; Zylicz, A.; Walerych, D.; Kampinga, H.H. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 2011, 435, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Hyder, I.; Pasumarti, M.; Reddy, P.R.; Prasad, C.S.; Kumar, K.A.; Sejian, V. Thermotolerance in Domestic Ruminants: A HSP70 Perspective. In Heat Shock Proteins and Stress; Springer: Singapore, 2017; Volume 12, pp. 3–35. [Google Scholar]
- Santpere, G.; Esparcia, P.G.; Benito, P.A.; Lorente-Galdos, B.; Navarro, A.; Ferrer, I. Transcriptional network analysis in frontal cortex in Lewy body diseases with focus on dementia with Lewy bodies. Brain Pathol. 2018, 28, 315–333. [Google Scholar] [CrossRef]
- Henderson-Smith, A.; Corneveaux, J.J.; De Both, M.; Cuyugan, L.; Liang, W.S.; Huentelman, M.; Adler, C.; Driver-Dunckley, E.; Beach, T.G.; Dunckley, T.L. Next-generation profiling to identify the molecular etiology of Parkinson dementia. Neurol. Genet. 2016, 2, e75. [Google Scholar] [CrossRef] [PubMed]
- Annese, A.; Manzari, C.; Lionetti, C.; Picardi, E.; Horner, D.S.; Chiara, M.; Caratozzolo, M.F.; Tullo, A.; Fosso, B.; Pesole, G.; et al. Whole transcriptome profiling of Late-Onset Alzheimer’s Disease patients provides insights into the molecular changes involved in the disease. Sci. Rep. 2018, 8, 4282. [Google Scholar] [CrossRef]
- Becirovic, L.; Brown, I.R. Targeting of Heat Shock Protein HSPA6 (HSP70B′) to the Periphery of Nuclear Speckles is Disrupted by a Transcription Inhibitor Following Thermal Stress in Human Neuronal Cells. Neurochem. Res. 2016, 42, 406–414. [Google Scholar] [CrossRef]
- Deane, C.; Brown, I.R. Differential Targeting of Hsp70 Heat Shock Proteins HSPA6 and HSPA1A with Components of a Protein Disaggregation/Refolding Machine in Differentiated Human Neuronal Cells following Thermal Stress. Front. Neurosci. 2017, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Deane, C.; Brown, I.R. Knockdown of Heat Shock Proteins HSPA6 (Hsp70B’) and HSPA1A (Hsp70-1) Sensitizes Differentiated Human Neuronal Cells to Cellular Stress. Neurochem. Res. 2017, 43, 340–350. [Google Scholar] [CrossRef]
- Su, Y.-S.; Hwang, L.-H.; Chen, C.-J. Heat Shock Protein A6, a Novel HSP70, Is Induced During Enterovirus A71 Infection to Facilitate Internal Ribosomal Entry Site-Mediated Translation. Front. Microbiol. 2021, 12, 664955. [Google Scholar] [CrossRef]
- Wang, L.; Hou, J.; Wang, J.; Zhu, Z.; Zhang, W.; Zhang, X.; Shen, H.; Wang, X. Regulatory roles of HSPA6 in Actinidia chinensis Planch. root extract (acRoots)-inhibited lung cancer proliferation. Clin. Transl. Med. 2020, 10, e46. [Google Scholar] [CrossRef]
- Shen, S.; Wei, C.; Fu, J. RNA-Sequencing Reveals Heat Shock 70-kDa Protein 6 (HSPA6) as a Novel Thymoquinone-Upregulated Gene That Inhibits Growth, Migration, and Invasion of Triple-Negative Breast Cancer Cells. Front. Oncol. 2021, 11, 667995. [Google Scholar] [CrossRef]
- Shin, S.-S.; Song, J.-H.; Hwang, B.; Noh, D.-H.; Park, S.L.; Kim, W.T.; Kim, W.-J.; Moon, S.-K. HSPA6 augments garlic extract-induced inhibition of proliferation, migration, and invasion of bladder cancer EJ cells; Implication for cell cycle dysregulation, signaling pathway alteration, and transcription factor-associated MMP-9 regulation. PLoS ONE 2017, 12, e0171860. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.C.; Castro, M.G.; Maenza, J.; Weis, J.P.; Kwong, G.A. Remote Control of Mammalian Cells with Heat-Triggered Gene Switches and Photothermal Pulse Trains. ACS Synth. Biol. 2018, 7, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Sun, R.; Qi, C.; Jiao, S.; Jiang, Y.; Liu, Z.; Zhao, D.; Liu, R.; Li, Q.; Yang, K.; et al. Generation of a pHSPA6 gene-based multifunctional live cell sensor. Biochim. Biophys. Acta (BBA) Bioenerg. 2021, 1868, 118919. [Google Scholar] [CrossRef]
- Leung, T.K.; Rajendran, M.Y.; Monfries, C.; Hall, C.; Lim, L. The human heat-shock protein family. Expression of a novel heat-inducible HSP70 (HSP70B′) and isolation of its cDNA and genomic DNA. Biochem. J. 1990, 267, 125–132. [Google Scholar] [CrossRef]
- Dezeure, F.; Vaiman, M.; Chardon, P. Characterization of a polymorphic heat shock protein 70 gene in swine outside the SLA major histocompatibility complex. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1993, 1174, 17–26. [Google Scholar] [CrossRef]
- Parsian, A.J.; Sheren, J.E.; Tao, T.Y.; Goswami, P.; Malyapa, R.; Van Rheeden, R.; Watson, M.S.; Hunt, C.R. The human Hsp70B gene at the HSPA7 locus of chromosome 1 is transcribed but non-functional. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 2000, 1494, 201–205. [Google Scholar] [CrossRef]
- Hatayama, T.; Asai, Y.; Wakatsuki, T.; Kitamura, T.; Imahara, H. Regulation of hsp70 Synthesis Induced by Cupric Sulfate and Zinc Sulfate in Thermotolerant HeLa Cells. J. Biochem. 1993, 114, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.R.; Hanson, P.J. Induction of heat shock protein 72 by a nitric oxide donor in guinea-pig gastric mucosal cells. Eur. J. Pharmacol. 1998, 353, 117–122. [Google Scholar] [CrossRef]
- Xu, Q.; Hu, Y.; Kleindienst, R.; Wick, G. Nitric oxide induces heat-shock protein 70 expression in vascular smooth muscle cells via activation of heat shock factor 1. J. Clin. Investig. 1997, 100, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Levinson, W.; Oppermann, H.; Jackson, J. Transition series metals and sulfhydryl reagents induce the synthesis of four proteins in eukaryotic cells. Biochim. Biophys. Acta (BBA) Nucleic Acids Protein Synth. 1980, 606, 170–180. [Google Scholar] [CrossRef]
- Romano, C.; Benedetto, N.; Catania, M.; Rizzo, A.; Gallè, F.; Losi, E.; Hasty, D.; Rossano, F. Commonly used antibiotics induce expression of Hsp 27 and Hsp 60 and protect human lymphocytes from apoptosis. Int. Immunopharmacol. 2004, 4, 1067–1073. [Google Scholar] [CrossRef]
- Rohmer, S.; Mainka, A.; Knippertz, I.; Hesse, A.; Nettelbeck, D.M. Insulated hsp70B′ promoter: Stringent heat-inducible activity in replication-deficient, but not replication-competent adenoviruses. J. Gene Med. 2008, 10, 340–354. [Google Scholar] [CrossRef]
- Wada, K.-I.; Taniguchi, A.; Okano, T. Highly sensitive detection of cytotoxicity using a modified HSP70B′ promoter. Biotechnol. Bioeng. 2007, 97, 871–876. [Google Scholar] [CrossRef]
- Ramirez, V.P.; Stamatis, M.; Shmukler, A.; Aneskievich, B.J. Basal and stress-inducible expression of HSPA6 in human keratinocytes is regulated by negative and positive promoter regions. Cell Stress Chaperon. 2014, 20, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Chow, A.M.; Mok, P.; Xiao, D.; Khalouei, S.; Brown, I.R. Heteromeric complexes of heat shock protein 70 (HSP70) family members, including Hsp70B′, in differentiated human neuronal cells. Cell Stress Chaperon. 2010, 15, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Noonan, E.J.; Place, R.F.; Rasoulpour, R.J.; Giardina, C.; Hightower, L.E. Cell number-dependent regulation of Hsp70B′ expression: Evidence of an extracellular regulator. J. Cell. Physiol. 2007, 210, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.M.; Larin, Z.; Taylor, I.C.; Prentice, H.; A Gwinn, K.; E Kingston, R. Multiple basal elements of a human hsp70 promoter function differently in human and rodent cell lines. Mol. Cell. Biol. 1987, 7, 3646–3655. [Google Scholar] [CrossRef]
- Giardina, C.; Pérez-Riba, M.; Lis, J.T. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992, 6, 2190–2200. [Google Scholar] [CrossRef]
- Lis, J.; Wu, C. Protein traffic on the heat shock promoter: Parking, stalling, and trucking along. Cell 1993, 74, 1–4. [Google Scholar] [CrossRef]
- Tang, H.; Liu, Y.; Madabusi, L.; Gilmour, D.S. Promoter-Proximal Pausing on the hsp70 Promoter in Drosophila melanogaster Depends on the Upstream Regulator. Mol. Cell. Biol. 2000, 20, 2569–2580. [Google Scholar] [CrossRef]
- Deane, C.A.; Brown, I.R. Intracellular Targeting of Heat Shock Proteins in Differentiated Human Neuronal Cells Following Proteotoxic Stress. J. Alzheimer’s Dis. 2018, 66, 1295–1308. [Google Scholar] [CrossRef]
- Tukaj, S. Heat Shock Protein 70 as a Double Agent Acting Inside and Outside the Cell: Insights into Autoimmunity. Int. J. Mol. Sci. 2020, 21, 5298. [Google Scholar] [CrossRef]
- Evans, C.G.; Chang, C.Y.; Gestwicki, J.E. Heat Shock Protein 70 (Hsp70) as an Emerging Drug Target. J. Med. Chem. 2010, 53, 4585–4602. [Google Scholar] [CrossRef] [PubMed]
- Nitika; Blackman, J.S.; Knighton, L.E.; Takakuwa, J.E.; Calderwood, S.K.; Truman, A.W. Chemogenomic screening identifies the Hsp70 co-chaperone DNAJA1 as a hub for anticancer drug resistance. Sci. Rep. 2020, 10, 13831. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.A.; Kim, Y.S.; Rivera-Marquez, G.M.; Oshima, N.; Watson, M.J.; Beebe, K.E.; Wells, C.; Lee, S.; Zuehlke, A.D.; Shao, H.; et al. Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer. Cancer Res. 2018, 78, 4022–4035. [Google Scholar] [CrossRef]
- Tukaj, S.; Węgrzyn, G. Anti-Hsp90 therapy in autoimmune and inflammatory diseases: A review of preclinical studies. Cell Stress Chaperon. 2016, 21, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Zhou, J.; Lu, Y.; Guan, L.; Wei, N.-N.; Tang, Y.-Q.; Wang, K. Inhibition of Hsp70 Suppresses Neuronal Hyperexcitability and Attenuates Epilepsy by Enhancing A-Type Potassium Current. Cell Rep. 2019, 26, 168–181.e4. [Google Scholar] [CrossRef]
- Luo, W.; Sun, W.; Taldone, T.; Rodina, A.; Chiosis, G. Heat shock protein 90 in neurodegenerative diseases. Mol. Neurodegener. 2010, 5, 24. [Google Scholar] [CrossRef]
- Kuballa, P.; Baumann, A.-L.; Mayer, K.; Bär, U.; Burtscher, H.; Brinkmann, U. Induction of heat shock protein HSPA6 (HSP70B′) upon HSP90 inhibition in cancer cell lines. FEBS Lett. 2015, 589, 1450–1458. [Google Scholar] [CrossRef]
- Badri, T.M.; Chen, K.L.; Alsiddig, M.A.; Li, L.; Cai, Y.; Wang, G.L. Genetic polymorphism in Hsp90AA1 gene is associated with the thermotolerance in Chinese Holstein cows. Cell Stress Chaperon. 2018, 23, 639–651. [Google Scholar] [CrossRef]
- Hassan, F.; Nawaz, A.; Rehman, M.S.; Ali, M.A.; Dilshad, S.M.; Yang, C. Prospects of HSP70 as a genetic marker for thermo-tolerance and immuno-modulation in animals under climate change scenario. Anim. Nutr. 2019, 5, 340–350. [Google Scholar] [CrossRef]

| Name | Primer | Sequence (5′–3′) |
|---|---|---|
| sgRNA-H1 | sgRNA-H1-F | CACCGTTCGCCCGGAAGGCGCTGGA |
| sgRNA-H1-R | AAACTCCAGCGCCTTCCGGGCGAAC | |
| sgRNA-H2 | sgRNA-H2-F | CACCGTAGAACCAGCTGGGAGAAGC |
| sgRNA-H2-R | AAACGCTTCTCCCAGCTGGTTCTAC | |
| sgRNA1-1 | sgRNA1-1-F | CACCGATTGTCACAAATCCTAGTAG |
| sgRNA1-1-R | AAACCTACTAGGATTTGTGACAATC | |
| sgRNA1-2 | sgRNA1-2-F | CACCGTTAACATCATCCCTAGCCCT |
| sgRNA1-2-R | AAACAGGGCTAGGGATGATGTTAAC |
| Name | Primer | Sequence (5′–3′) | Usage |
|---|---|---|---|
| HSE-1-KO | H1-F | CTCTCTTCCCATGGTGA | PCR |
| H1-R | GCTGGTGCATCTGACTTCAT | ||
| HSE-2-KO | H2-F | CTCTCTTCCCATGGTGA | PCR |
| H2-R | GCTGGTGCATCTGACTTCAT | ||
| CR-KO | F1-F | CCTTTCTGGGCTGCGACTTGAT | PCR |
| R1-R | GGGCGGATCGTCTGTTCAAGGA | ||
| EGFP-KI [21] | EGFP-KI -F | GAGGCGCATGTTCTCCAAAAACC | PCR |
| EGFP-KI -R | AGCCACACTTGTAGTTGCACTGG | ||
| pHSPA6 [21] | pHSPA6 q-F | ATCCATGATATTGTCCTA | qPCR |
| pHSPA6 q-R | TTATGCTCTTGTTCAGTT | ||
| GS [21] | GS q-F | CTTGCATCGTGTGTGCGAAG | qPCR |
| GS q-R | GCTTAGCTTCTCGATGGCCT | ||
| DNAJB1 [21] | B1 q-F | TGACCATCGAAGTGAAGCGG | qPCR |
| B1 q-R | TCGGCTGGAATGTTGTTGGA | ||
| DNAJC3 [21] | C3 q-F | GGAGCCTGACAATGTGAATGC | qPCR |
| C3 q-R | GACCTTCTCGAATCTGCTGGT | ||
| pHSP70.2 | 70 q-F | GAGCAAGGAGGAGATCGAGC | qPCR |
| 70 q-R | GTTGAAGGCGTACGACTCCA | ||
| pHSP90AA1 | 90 q-F | TCGAAGGGCAGTTGGAGTTC | qPCR |
| 90 q-R | ATGAGCTCCTCGCAGTTGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, S.; Bai, C.; Qi, C.; Wu, H.; Hu, L.; Li, F.; Yang, K.; Zhao, C.; Ouyang, H.; Pang, D.; et al. Identification and Functional Analysis of the Regulatory Elements in the pHSPA6 Promoter. Genes 2022, 13, 189. https://doi.org/10.3390/genes13020189
Jiao S, Bai C, Qi C, Wu H, Hu L, Li F, Yang K, Zhao C, Ouyang H, Pang D, et al. Identification and Functional Analysis of the Regulatory Elements in the pHSPA6 Promoter. Genes. 2022; 13(2):189. https://doi.org/10.3390/genes13020189
Chicago/Turabian StyleJiao, Shuyu, Chunyan Bai, Chunyun Qi, Heyong Wu, Lanxin Hu, Feng Li, Kang Yang, Chuheng Zhao, Hongsheng Ouyang, Daxin Pang, and et al. 2022. "Identification and Functional Analysis of the Regulatory Elements in the pHSPA6 Promoter" Genes 13, no. 2: 189. https://doi.org/10.3390/genes13020189
APA StyleJiao, S., Bai, C., Qi, C., Wu, H., Hu, L., Li, F., Yang, K., Zhao, C., Ouyang, H., Pang, D., Tang, X., & Xie, Z. (2022). Identification and Functional Analysis of the Regulatory Elements in the pHSPA6 Promoter. Genes, 13(2), 189. https://doi.org/10.3390/genes13020189






