Prenatal Diagnosis of Placental Mesenchymal Dysplasia with 46, X, Isochromosome Xq/45, X Mosaicism
Abstract
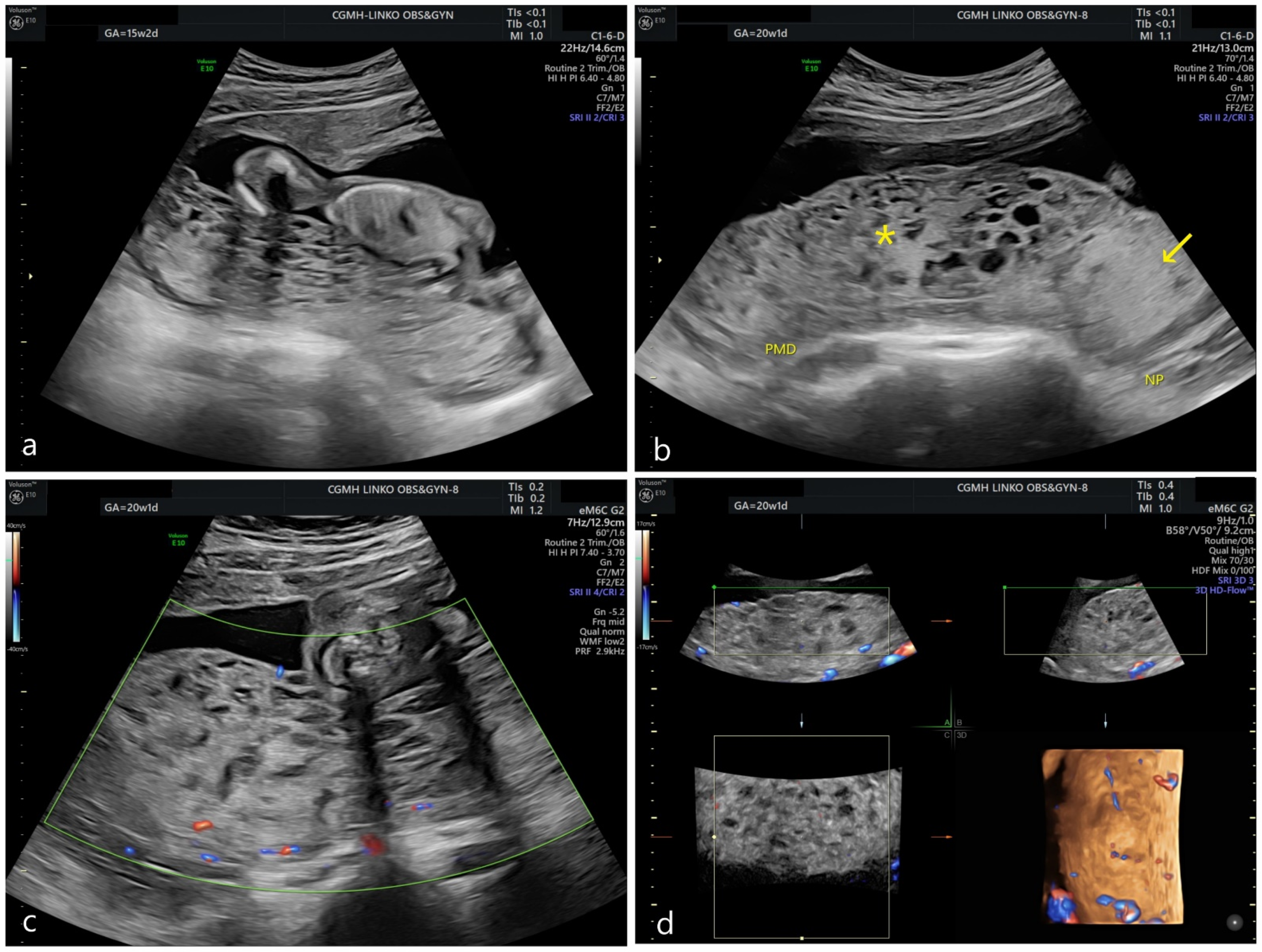
:1. Introduction
2. Materials and Methods
2.1. Patient
2.2. Cytogenetic Studies: Cell Culture and Karyotyping
2.3. DNA Extraction
2.4. Array Comparative Genomic Hybridization (Array CGH)
2.5. Histology
2.6. Short Tandem Repeat (STR) Analysis
2.7. Methylation-Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA)
2.8. Mosaic Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hodgson, A.; Dube, V.; Strickland, S.; Kolomietz, E.; Noor, A.; Akbari, A.; Colgan, T.; Turashvili, G. Androgenetic/Biparental Mosaic/Chimeric Conceptions with a Molar Component: A Diagnostic and Clinical Challenge. Int. J. Gynecol. Pathol. 2021, 40, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Nayeri, U.A.; West, A.B.; Grossetta Nardini, H.K.; Copel, J.A.; Sfakianaki, A.K. Systematic review of sonographic findings of placental mesenchymal dysplasia and subsequent pregnancy outcome. Ultrasound Obstet. Gynecol. 2013, 41, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Kaiser-Rogers, K.A.; McFadden, D.E.; Livasy, C.A.; Dansereau, J.; Jiang, R.; Knops, J.F.; Lefebvre, L.; Rao, K.W.; Robinson, W.P. Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J. Med. Genet. 2006, 43, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benn, P.A.; Clive, J.M.; Collins, R. Medians for second-trimester maternal serum alpha-fetoprotein, human chorionic gonadotropin, and unconjugated estriol; differences between races or ethnic groups. Clin. Chem. 1997, 43, 333–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwa, H.L.; Chang, Y.Y.; Lee, J.C.; Yin, H.Y.; Chen, Y.H.; Tseng, L.H.; Su, Y.N.; Ko, T.M. Thirteen X-chromosomal short tandem repeat loci multiplex data from Taiwanese. Int. J. Leg. Med. 2009, 123, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Allias, F.; Lebreton, F.; Collardeau-Frachon, S.; Azziza, J.; Pasquier, C.J.; Arcin-Thoury, F.; Patrier, S.; Devouassoux-Shisheboran, M. Immunohistochemical expression of p57 in placental vascular proliferative disorders of preterm and term placentas. Fetal Pediatr. Pathol. 2009, 28, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Chen, M.F.; Bureau, Y.A.; Brown, R. Placental mesenchymal dysplasia and an estimation of the population incidence. Acta Obstet. Gynecol. Scand. 2012, 91, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Arizawa, M.; Nakayama, M. Suspected involvement of the X chromosome in placental mesenchymal dysplasia. Congenit. Anom. (Kyoto) 2002, 42, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Guzel Erdal, G.; Balkan, M. Turner Syndrome with Isochromosome Xq. J. Pediatr. Genet. 2020, 9, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.S.; Sandstrom, M.; Goldstein, D.P.; Driscoll, S.G. 45,X complete hydatidiform mole. Gynecol. Oncol. 1982, 14, 279–283. [Google Scholar] [CrossRef]
- Destouni, A.; Zamani Esteki, M.; Catteeuw, M.; Tšuiko, O.; Dimitriadou, E.; Smits, K.; Kurg, A.; Salumets, A.; Van Soom, A.; Voet, T.; et al. Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res. 2016, 26, 567–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callen, D.F.; Mulley, J.C.; Baker, E.G.; Sutherland, G.R. Determining the origin of human X isochromosomes by use of DNA sequence polymorphisms and detection of an apparent i(Xq) with Xp sequences. Hum. Genet. 1987, 77, 236–240. [Google Scholar] [CrossRef]





| Amniotic Fluid | Cord Blood | Non-Dysplastic Placenta | Dysplastic Placenta | |
|---|---|---|---|---|
| Mean log ratio | 0.591397 | 0.572296 | 0.251074 | −0.46537 |
| N/2 | 1.506705 | 1.486888 | 1.190093 | 0.724287 |
| N | 3.01341 | 2.973776 | 2.380186 | 1.448574 |
| δ | 100.67% | 98.69% | 69.01% | 22.43% |
| 1−δ | −0.67% | 1.31% | 30.99% | 77.57% |
| Amniotic Fluid | Cord Blood | Non-Dysplastic Placenta | Dysplastic Placenta | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STR Markers | Chromosomal Location | Allele (P/M) | AUC (P/M) | Biparental Rate | Allele (P/M) | AUC (P/M) | Biparental Rate | Allele (P/M) | AUC (P/M) | Biparental Rate | Allele (P/M) | AUC (P/M) | Biparental Rate |
| D3S1358 | 3p21.31 | 14/17 | 16,751/14,151 | 91.59% | 14/17 | 18,393/17,785 | 98.32% | 14/17 | 15,856/14,804 | 96.57% | 14/17 | 48,076/6164 | 22.73% |
| vWA | 12p13.31 | 17/18 | 27,518/24,229 | 93.64% | 17/18 | 41,960/33,329 | 88.54% | 17/18 | 27,448/30,097 | 104.60% | 17/18 | 66,220/11,697 | 30.02% |
| D16S539 | 16q24.1 | 9/11 | 32,759/36,536 | 105.45% | 9/11 | 78,624/76,666 | 98.74% | 9/11 | 66,014/61,394 | 96.37% | 9/11 | 120,006/11,389 | 17.34% |
| D8S1179 | 8q24.13 | 11/14 | 16,840/11,381 | 80.66% | 11/14 | 18,445/22,999 | 110.99% | 11/14 | 18,056/16,524 | 95.57% | 11/14 | 48,812/6321 | 22.93% |
| D18S51 | 21q11.2–q21 | 15/16 | 93,826/104,530 | 105.40% | 15/16 | 149,449/118,218 | 88.33% | 15/16 | 102,685/98,713 | 98.03% | 15/16 | 158,985/31,591 | 33.15% |
| D2S441 | 18q21.33 | 11/13 | 39,236/27,800 | 82.94% | 11/13 | 47,803/47,727 | 99.92% | 11/13 | 41,749/43,326 | 101.85% | 11/13 | 86,278/14,227 | 28.31% |
| TH01 | 2p14 | 9/7 | 42,235/53,801 | 112.04% | 9/7 | 70,117/80,395 | 106.83% | 9/7 | 63,412/66,071 | 102.05% | 9/7 | 109,520/18,859 | 29.38% |
| FGA | 19q12 | 20/23 | 45,652/55,661 | 109.88% | 20/23 | 97,451/96,252 | 99.38% | 20/23 | 69,714/83,480 | 108.99% | 20/23 | 111,815/14,095 | 22.39% |
| D5S818 | 11p15.5 | 9/13 | 36,069/24,468 | 80.84% | 9/13 | 38,269/37,060 | 98.40% | 9/13 | 25,995/25,416 | 98.87% | 9/13 | 78,297/10,763 | 24.17% |
| D13S317 | 4q28 | 10/11 | 42,997/42,240 | 99.11% | 10/11 | 70,928/54,565 | 86.96% | 10/11 | 37,985/39,182 | 101.55% | 10/11 | 95,033/13,547 | 24.95% |
| SE33 | 5q21–31 | 27.2/18 | 128,927/130,009 | 100.42% | 27.2/18 | 213,111/210,357 | 99.35% | 27.2/18 | 180,405/160,701 | 94.22% | 27.2/18 | 218,007/48,467 | 36.38% |
| D10S1248 | 13q22–31 | 14/15 | 18,771/14,955 | 88.69% | 14/15 | 18,719/19,616 | 102.34% | 14/15 | 13,096/11,450 | 93.29% | 14/15 | 49,175/3700 | 14.00% |
| D12S391 | 6q14 | 23/18 | 40,624/42,025 | 101.70% | 23/18 | 55,074/72,839 | 113.89% | 23/18 | 37,519/42,589 | 106.33% | 23/18 | 98,544/16,879 | 29.25% |
| D2S1338 | 10q26.3 | 19/24 | 67,167/65,573 | 98.80% | 19/24 | 69,352/61,456 | 93.96% | 19/24 | 63,880/65,648 | 101.36% | 19/24 | 159,569/23,995 | 26.14% |
| Average | 96.51% | Average | 99.00% | Average | 99.98% | Average | 25.80% | ||||||
| Amniotic Fluid | Cord Blood | Non-Dysplastic Placenta | Dysplastic Placenta | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STR Markers | Chromo-Somal Location | Allele (P/M) | AUC (P/M) | 46, X, isoXq Rate | Allele (P/M) | AUC (P/M) | 46, X, isoXq Rate | Allele (P/M) | AUC (P/M) | 46, X, isoXq Rate | Allele (P/M) | AUC (P/M) | 46, X, isoXq Rate |
| DXS6807 | Xp | 11/x(15) | 12,848/0 | 11/x(15) | 55,158/0 | 11/x(15) | 49,061/0 | 11/x(15) | 97,868/0 | ||||
| DXS9902 | Xp | 10/x(11) | 20,816/0 | 10/x(11) | 47,862/0 | 10/x(11) | 51,007/0 | 10/x(11) | 83,574/0 | ||||
| DXS9898 | Xq | 14/12 | 15,843/32,626 | 102.97% | 14/12 | 60,309/126,386 | 104.78% | 14/12 | 54,856/81,814 | 74.57% | 14/12 | 112,201/11,273 | 5.02% |
| DXS8377 | Xq | 48/51 | 25,374/49,396 | 97.34% | 48/51 | 65,812/134,888 | 102.48% | 48/51 | 68,455/83,375 | 60.90% | 48/51 | 142,370/17,128 | 6.02% |
| HPRTB | Xq | 15/12 | 28,132/53,429 | 94.96% | 15/12 | 62,571/149,412 | 119.39% | 15/12 | 71,063/100,667 | 70.83% | 15/12 | 145,559/23,456 | 8.06% |
| DXS7132 | Xq | 14/15 | 36,437/60,358 | 82.83% | 14/15 | 119,612/162,804 | 68.06% | 14/15 | 88,222/112,703 | 63.87% | 14/15 | 165,957/26,683 | 8.04% |
| DXS101 | Xq | 28/26 | 17,510/37,195 | 106.21% | 28/26 | 47,461/92,150 | 97.08% | 28/26 | 41,584/65,929 | 79.27% | 28/26 | 93,974/20,712 | 11.02% |
| DXS6789 | Xq | 15/21 | 69,262/142,895 | 103.16% | 15/21 | 175,639/22,9088 | 65.22% | 15/21 | 164,974/190,858 | 57.84% | 15/21 | 245,485/43,987 | 8.96% |
| DXS7424 | Xq | 15/16 | 18,594/25,692 | 69.09% | 15/16 | 38,859/61,909 | 79.66% | 15/16 | 43,348/42,893 | 49.48% | 15/16 | 66,848/13,377 | 10.01% |
| Average | 93.79% | Average | 90.95% | Average | 65.25% | Average | 8.16% | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, C.-C.; Lee, C.-H.; Chang, S.-D.; Ko, T.-M.; Ueng, S.-H.; Chen, Y.-H.; Wang, M.-C.; Chang, Y.-L. Prenatal Diagnosis of Placental Mesenchymal Dysplasia with 46, X, Isochromosome Xq/45, X Mosaicism. Genes 2022, 13, 245. https://doi.org/10.3390/genes13020245
Hsu C-C, Lee C-H, Chang S-D, Ko T-M, Ueng S-H, Chen Y-H, Wang M-C, Chang Y-L. Prenatal Diagnosis of Placental Mesenchymal Dysplasia with 46, X, Isochromosome Xq/45, X Mosaicism. Genes. 2022; 13(2):245. https://doi.org/10.3390/genes13020245
Chicago/Turabian StyleHsu, Chin-Chieh, Chien-Hong Lee, Shuenn-Dyh Chang, Tsang-Ming Ko, Shir-Hwa Ueng, Yu-Hsiu Chen, Mei-Chia Wang, and Yao-Lung Chang. 2022. "Prenatal Diagnosis of Placental Mesenchymal Dysplasia with 46, X, Isochromosome Xq/45, X Mosaicism" Genes 13, no. 2: 245. https://doi.org/10.3390/genes13020245
APA StyleHsu, C.-C., Lee, C.-H., Chang, S.-D., Ko, T.-M., Ueng, S.-H., Chen, Y.-H., Wang, M.-C., & Chang, Y.-L. (2022). Prenatal Diagnosis of Placental Mesenchymal Dysplasia with 46, X, Isochromosome Xq/45, X Mosaicism. Genes, 13(2), 245. https://doi.org/10.3390/genes13020245






