The Ribosomal Protein RpL22 Interacts In Vitro with 5′-UTR Sequences Found in Some Drosophila melanogaster Transposons
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Analysis
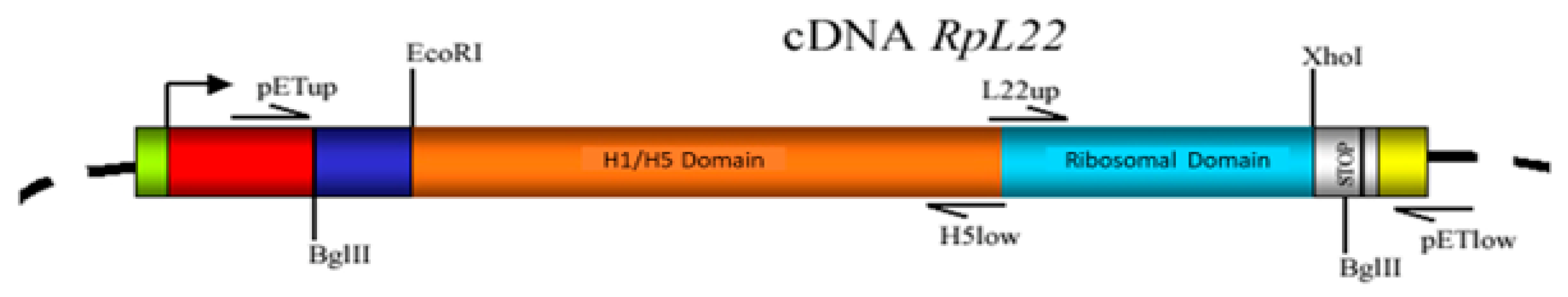
2.2. Plasmid Construction and Sequencing
2.3. Yeast One-Hybrid Assay
2.4. Expression and Purification of RpL22 Protein and RpL22/H5 RpL22/L22 Polypeptides
2.5. DNA-Binding Assays
2.6. Production of Antibody Anti RpL22/H5
2.7. Immunofluorescence and Immunocytochemistry
3. Results
3.1. Search for Shared Motifs
3.2. Identification of Proteins Able to Interact with TERM
3.3. Analysis of RpL22/TERM Interaction
3.4. RpL22 Sub-Cellular Localization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McClintock, B. Induction of Instability at Selected Loci in Maize. Genetics 1953, 38, 579–599. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [Green Version]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dahlstrom, J.E.; Chandra, A.; Board, P.; Rangasamy, D. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res. Treat. 2012, 136, 129–142. [Google Scholar] [CrossRef] [Green Version]
- Lannoy, N.; Hermans, C. Principles of genetic variations and molecular diseases: Applications in hemophilia A. Crit. Rev. Oncol./Hematol. 2016, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jin, Y.; Prazak, L.; Hammell, M.; Dubnau, J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE 2012, 7, e44099. [Google Scholar] [CrossRef] [Green Version]
- Solyom, S.; Ewing, A.D.; Rahrmann, E.P.; Doucet, T.; Nelson, H.H.; Burns, M.B.; Harris, R.S.; Sigmon, D.F.; Casella, A.; Erlanger, B.; et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012, 22, 2328–2338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, M.T.; Faulkner, G.J.; Dubnau, J.; Ponomarev, I.; Gage, F.H. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 2013, 33, 17577–17586. [Google Scholar] [CrossRef] [Green Version]
- Barrón, M.G.; Fiston-Lavier, A.-S.; Petrov, D.A.; González, J. Population Genomics of Transposable Elements in Drosophila. Annu. Rev. Genet. 2014, 48, 561–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminker, J.S.; Bergman, C.M.; Kronmiller, B.; Carlson, J.; Svirskas, R.; Patel, S.; Frise, E.; Wheeler, D.A.; Lewis, S.E.; Rubin, G.M.; et al. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 2002, 3, RESEARCH0084. [Google Scholar] [CrossRef] [Green Version]
- Boeke, J.D.; Garfinkel, D.J.; Styles, C.A.; Fink, G.R. Ty elements transpose through an RNA intermediate. Cell 1985, 40, 491–500. [Google Scholar] [CrossRef]
- McCullers, T.J.; Steiniger, M. Transposable elements in Drosophila. Mob. Genet. Elem. 2017, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Venner, S.; Feschotte, C.; Biemont, C. Dynamics of transposable elements: Towards a community ecology of the genome. Trends Genet. 2009, 25, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, K.; Largaespada, D.A.; Ivics, Z. Transposons As Tools for Functional Genomics in Vertebrate Models. Trends Genet. 2017, 33, 784–801. [Google Scholar] [CrossRef]
- Palazzo, A.; Marsano, R.M. Transposable elements: A jump toward the future of expression vectors. Crit. Rev. Biotechnol. 2021, 1–27. [Google Scholar] [CrossRef]
- Minervini, C.F.; Ruggieri, S.; Traversa, M.; D’Aiuto, L.; Marsano, R.M.; Leronni, D.; Centomani, I.; De Giovanni, C.; Viggiano, L. Evidences for insulator activity of the 5′UTR of the Drosophila melanogaster LTR-retrotransposon ZAM. Mol. Genet. Genom. 2010, 283, 503–509. [Google Scholar] [CrossRef]
- Minervini, C.F.; Marsano, R.M.; Casieri, P.; Fanti, L.; Caizzi, R.; Pimpinelli, S.; Rocchi, M.; Viggiano, L. Heterochromatin protein 1 interacts with 5′UTR of transposable element ZAM in a sequence-specific fashion. Gene 2007, 393, 1–10. [Google Scholar] [CrossRef]
- Viggiano, L.; Caggese, C.; Barsanti, P.; Caizzi, R. Cloning and characterization of a copy of Tirant transposable element in Drosophila melanogaster. Gene 1997, 197, 29–35. [Google Scholar] [CrossRef]
- Marsano, R.M.; Moschetti, R.; Caggese, C.; Lanave, C.; Barsanti, P.; Caizzi, R. The complete Tirant transposable element in Drosophila melanogaster shows a structural relationship with retrovirus-like retrotransposons. Gene 2000, 247, 87–95. [Google Scholar] [CrossRef]
- Desset, S.; Conte, C.; Dimitri, P.; Calco, V.; Dastugue, B.; Vaury, C. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol. 1999, 16, 54–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Bidwai, A.P.; Glover, C.V.C. Interaction of casein kinase II with ribosomal protein L22 of Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2002, 298, 60–66. [Google Scholar] [CrossRef]
- Koyama, Y.; Katagiri, S.; Hanai, S.; Uchida, K.; Miwa, M. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and L23a, with unique histone-like amino-terminal extensions. Gene 1999, 226, 339–345. [Google Scholar] [CrossRef] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Van Helden, J.; André, B.; Collado-Vides, J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies11Edited by G. von Heijne. J. Mol. Biol. 1998, 281, 827–842. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Turatsinze, J.-V.; Thomas-Chollier, M.; Defrance, M.; van Helden, J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008, 3, 1578–1588. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Garner, M.M.; Revzin, A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: Application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981, 9, 3047–3060. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, E.A.; DeCaprio, J.; Brahmandam, M. Detecting Protein Antigens in Sodium Dodecyl Sulfate-Polyacrylamide Gels. Cold Spring Harbor Protoc. 2019, 2019, pdb.prot099994. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, E.A.; DeCaprio, J.; Brahmandam, M. Preparing Protein Antigens from Sodium Dodecyl Sulfate-Polyacrylamide Gels for Immunization. Cold Spring Harbor Protoc. 2019, 2019, pdb.prot100008. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Mageeney, C.M.; Ware, V.C. Specialized eRpL22 paralogue-specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster. Mol. Biol. Cell 2019, 30, 2240–2253. [Google Scholar] [CrossRef]
- Chuong, E.B. The placenta goes viral: Retroviruses control gene expression in pregnancy. PLoS Biol. 2018, 16, e3000028. [Google Scholar] [CrossRef] [Green Version]
- Sundaram, V.; Wysocka, J. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190347. [Google Scholar] [CrossRef] [Green Version]
- Moschetti, R.; Palazzo, A.; Lorusso, P.; Viggiano, L.; Marsano, R.M. “What You Need, Baby, I Got It”: Transposable Elements as Suppliers of Cis-Operating Sequences in Drosophila. Biology 2020, 9, 25. [Google Scholar] [CrossRef] [Green Version]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [Green Version]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [Green Version]
- Mével-Ninio, M.; Pelisson, A.; Kinder, J.; Campos, A.R.; Bucheton, A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 2007, 175, 1615–1624. [Google Scholar] [CrossRef] [Green Version]
- Kassis, J.A. 14—Pairing-Sensitive Silencing, Polycomb Group Response Elements, and Transposon Homing in Drosophila. In Advertisment Genet; Dunlap, J.C., Wu, C.T., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 46, pp. 421–438. [Google Scholar]
- Georgiev, P.; Kozycina, M. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 1996, 142, 425–436. [Google Scholar] [CrossRef]
- Pai, C.Y.; Lei, E.P.; Ghosh, D.; Corces, V.G. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 2004, 16, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.-Q.; Liu, L.-P.; Hess, D.; Rietdorf, J.; Sun, F.-L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006, 20, 1959–1973. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.G.; Ireland, J.A.; Prem, S.M.; Chen, A.S.; Ware, V.C. RpL22e, but not RpL22e-like-PA, is SUMOylated and localizes to the nucleoplasm of Drosophila meiotic spermatocytes. Nucleus 2013, 4, 241–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berloco, M.F.; Minervini, C.F.; Moschetti, R.; Palazzo, A.; Viggiano, L.; Marsano, R.M. Evidence of the Physical Interaction between Rpl22 and the Transposable Element Doc5, a Heterochromatic Transposon of Drosophila melanogaster. Genes 2021, 12, 1997. [Google Scholar] [CrossRef] [PubMed]
- Arbeitman Michelle, N.; Furlong Eileen, E.M.; Imam, F.; Johnson, E.; Null Brian, H.; Baker Bruce, S.; Krasnow Mark, A.; Scott Matthew, P.; Davis Ronald, W.; White Kevin, P. Gene Expression During the Life Cycle of Drosophila melanogaster. Science 2002, 297, 2270–2275. [Google Scholar] [CrossRef]
- Lyne, R.; Smith, R.; Rutherford, K.; Wakeling, M.; Varley, A.; Guillier, F.; Janssens, H.; Ji, W.; McLaren, P.; North, P.; et al. FlyMine: An integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007, 8, R129. [Google Scholar] [CrossRef] [Green Version]
- Bischof, J.; Björklund, M.; Furger, E.; Schertel, C.; Taipale, J.; Basler, K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 2013, 140, 2434–2442. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Clone’s Name | BLAST Results | BLASTX Results | Notes |
|---|---|---|---|
| L1 | mitochondrial sequence | # | # |
| L2 | Grn | GRN | GATA trascription factor |
| L3 | CG7434 | RpL22 | Ribosomal protein |
| L4 | CG7434 | RpL22 | Ribosomal protein |
| L5 | Csn6 | CSN6 | Signalosome |
| L6 | CG7434 | RpL22 | Ribosomal protein |
| L7 | Mis | MIS | body pigmentation |
| pTERM3lig01 | CG4314 | st | eye pigment precursor transport |
| pTERM3lig02 | GS1 | GS1 | glutammina sintetasi |
| pTERM3lig03 | CG1883 | CG1883 | Rps7-like |
| pTERM3lig05 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig06 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig07 | Hrb27C | Hrb27C | RNA binding protein |
| pTERM3lig08 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig09 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig10 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig11 | CG9253 | CG9253 | RNA helicase activity |
| pTERM3lig12 | Mod(mdg4) | Mod(mdg4) | FLYWCH domain |
| pTERM3lig13 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig14 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig15 | RpS16 | RpS16 | Ribosomal protein |
| pTERM3lig16 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig17 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig19 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig20 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig21 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig25 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig26 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig27 | CG6007 | GatA | serine hydrolase activity |
| pTERM3lig29 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig31 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig32 | CG30389 | CG30389 | actin filament binding activity |
| pTERM3lig33 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig34 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig35 | CG9415 | CG9415 | trascription factor |
| pTERM3lig36 | CG9277 | CG9277 | beta tubulina |
| pTERM3lig38 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig39 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig41 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig43 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig44 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig45 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig46 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig47 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig48 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig49 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig50 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig51 | CG17326 | luna | Zinc finger C2H2-type |
| pTERM3lig52 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig53 | CG7434 | RpL22 | Ribosomal protein |
| pTERM3lig54 | CG7434 | RpL22 | Ribosomal protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minervini, C.F.; Berloco, M.F.; Marsano, R.M.; Viggiano, L. The Ribosomal Protein RpL22 Interacts In Vitro with 5′-UTR Sequences Found in Some Drosophila melanogaster Transposons. Genes 2022, 13, 305. https://doi.org/10.3390/genes13020305
Minervini CF, Berloco MF, Marsano RM, Viggiano L. The Ribosomal Protein RpL22 Interacts In Vitro with 5′-UTR Sequences Found in Some Drosophila melanogaster Transposons. Genes. 2022; 13(2):305. https://doi.org/10.3390/genes13020305
Chicago/Turabian StyleMinervini, Crescenzio Francesco, Maria Francesca Berloco, René Massimiliano Marsano, and Luigi Viggiano. 2022. "The Ribosomal Protein RpL22 Interacts In Vitro with 5′-UTR Sequences Found in Some Drosophila melanogaster Transposons" Genes 13, no. 2: 305. https://doi.org/10.3390/genes13020305






