Chloroplast Protein Tic55 Involved in Dark-Induced Senescence through AtbHLH/AtWRKY-ANAC003 Controlling Pathway of Arabidopsis thaliana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Genomic PCR and Semi-qRT-PCR
2.3. Stress Treatments
2.3.1. Heat Stress Treatment
2.3.2. Cold Stress Treatment
2.3.3. High Osmotic Pressure Treatment
2.4. Dark Treatment and Senescence-Related Gene Expression by Semi qRT-PCR
2.4.1. Dark-Induced Senescence
2.4.2. Semi-Quantitative RT-PCR Assays
2.4.3. Chlorophyll Concentration Analysis
2.5. Transactivation Assays in Yeast Cells
3. Results
3.1. Confirmation of tic55-II Knockout Mutant Line and Its Possible Biological Roles
3.2. Tic55 Involved in the Chlorophyll Metabolism during Plant Senescence
3.3. Possible TFs Regulate Plant Senescence
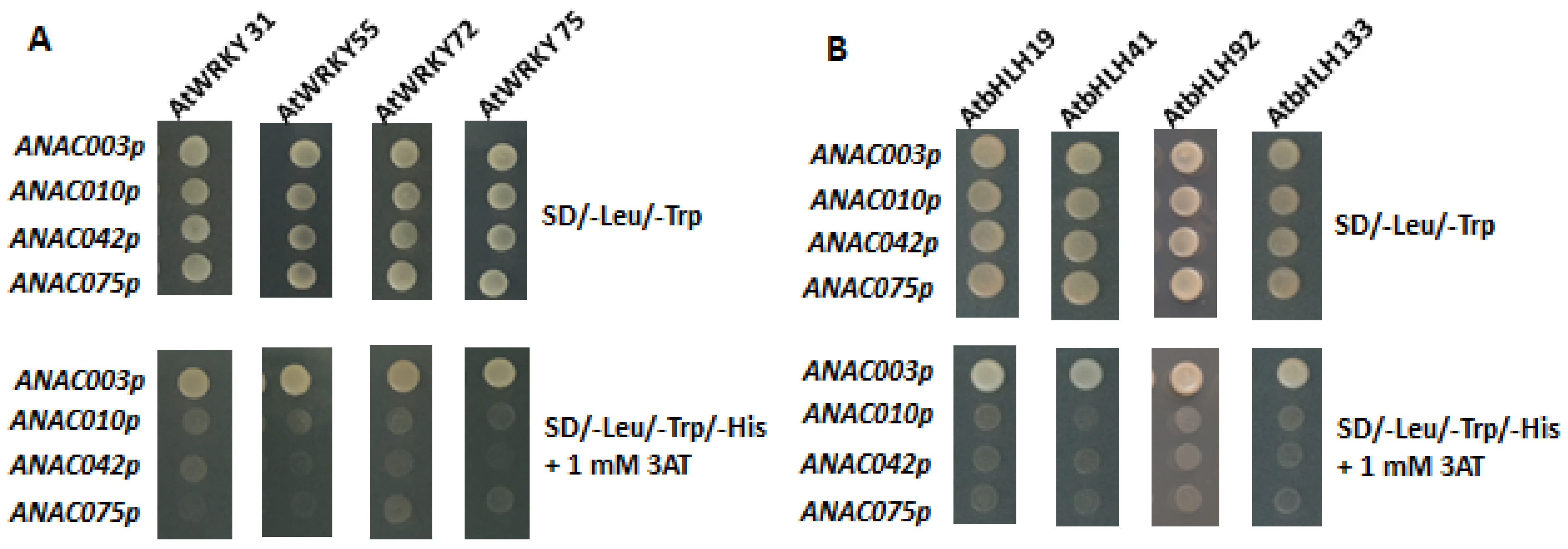
3.4. Yeast One-Hybrid Assays Showing Exoression of ANAC003 Is Regulated by AtWRKY and AtbHLH TFs, Respectively
3.5. Phylogenetic Analysis and Mutiple Sequence Alignment of AtWRKY and AtbHLH Proteins Related to Plant Senescence
3.6. Location of the Binding Region with Cis-Acting Elements in the ANAC003 Promoter for Binding with Different AtbHLH Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kubis, S.; Patel, R.; Combe, J.; Be’dard, J.; Kovacheva, S.; Lilley, K.; Biehl, A.; Leister, D.; Rı’os, G.; Koncz, C.; et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 2004, 16, 2059–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.M.; Chiu, C.C. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 2010, 61, 157–180. [Google Scholar] [CrossRef] [PubMed]
- Bölter, B.; Soll, J.; Schwenkert, S. Redox meets protein trafficking. Biochim. Biophys. Acta 2015, 1847, 949–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakai, M. The Tic complex uncovered: The alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta 2015, 1847, 957–967. [Google Scholar] [CrossRef] [Green Version]
- Chou, M.L.; Fitzpatric, L.M.; Tu, S.L.; Budziszewski, G.; Potter-Lewis, S.; Akita, M.; Levin, J.Z.; Keegstra, K.; Li, H.-M. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 2003, 22, 2970–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inaba, T.; Li, M.; Alvarez-Huerta, M.; Kessler, F.; Schnell, D.J. atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 2003, 278, 38617–38627. [Google Scholar] [CrossRef] [Green Version]
- Chou, M.L.; Chu, C.C.; Chen, L.J.; Akita, M.; Li, H.M. Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 2006, 175, 893–900. [Google Scholar] [CrossRef] [Green Version]
- Benz, J.P.; Soll, J.; Bölter, B. Protein transport in organelles: The composition, function and regulation of the Tic complex in chloroplast protein import. FEBS J. 2009, 276, 1166–1176. [Google Scholar] [CrossRef]
- Kikuchi, S.; Oishi, M.; Hirabayashi, Y.; Lee, D.W.; Hwang, I.; Nakai, M. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 2009, 21, 1781–1797. [Google Scholar] [CrossRef] [Green Version]
- Kovács-Bogdán, E.; Benz, J.P.; Soll, J.; Bölter, B. Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 2011, 11, 133–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, S.; Bedard, J.; Hirano, M.; Hirabayashi, Y.; Oishi, M.; Imai, M.; Takase, M.; Ide, T.; Nakai, M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 2013, 339, 571–574. [Google Scholar] [CrossRef]
- Balsera, M.; Goetze, T.A.; Kovacs-Bogdan, E.; Schurmann, P.; Wagner, R.; Buchanan, B.B.; Soll, J.; Bölter, B. Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J. Biol. Chem. 2009, 284, 2603–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stengel, A.; Benz, J.P.; Soll, J.; Bölter, B. Redox regulation of protein import into chloroplasts and mitochondria: Similarities and differences. Plant Signal. Behav. 2010, 5, 105–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stengel, A.; Benz, J.P.; Balsera, M.; Soll, J.; Bölter, B. Tic62 redox-regulated translocon composition and dynamics. J. Biol. Chem. 2008, 283, 6656–6667. [Google Scholar] [CrossRef] [Green Version]
- Chigri, F.; Hörmann, F.; Stamp, A.; Stammers, D.K.; Bölter, B.; Soll, J.; Vothknecht, U.C. Calcium regulation of chloroplast translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. USA 2006, 103, 16051–16056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hormann, F.; Kuchler, M.; Sveshnikov, D.; Oppermann, U.; Li, Y.; Soll, J. Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 2004, 279, 34756–34762. [Google Scholar] [CrossRef] [Green Version]
- Bartsch, S.; Monnet, J.; Selbach, K.; Quigley, F.; Gray, J.; von Wettstein, D.; Reinbothe, S.; Reinbothe, C. Three thioredoxin targets in the inner envelope membrane of chloroplasts function in protein import and chlorophyll metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 4933–4938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benz, J.P.; Stengel, A.; Lintala, M.; Lee, Y.H.; Weber, A.; Philippar, K.; Gügel, I.L.; Kaieda, S.; Ikegami, T.; Mulo, P.; et al. Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 2009, 21, 3965–3983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boij, P.; Patel, R.; Garcia, C.; Jarvis, P.; Aronsson, H. In vivo studies on the roles of Tic55-related proteins in chloroplast protein import in Arabidopsis thaliana. Mol. Plant 2009, 2, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Caliebe, A.; Grimm, R.; Kaiser, G.; Lubeck, J.; Soll, J.; Heins, L. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 1997, 16, 7342–7350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauenstein, M.; Christ, B.; Das, A.; Aubry, S.; Hörtensteiner, S. A Role for Tic55 as a hydroxylase of phyllobilins, the products of chlorophyll breakdown during plant senescence. Plant Cell 2016, 28, 2510–2527. [Google Scholar] [CrossRef] [Green Version]
- Chou, M.L.; Liao, W.Y.; Wei, W.C.; Li, A.Y.; Chu, C.Y.; Wu, C.L.; Liu, C.L.; Fu, T.H.; Lin, L.F. The direct involvement of dark-induced Tic55 protein in chlorophyll catabolism and its indirect role in the MYB108-NAC signaling pathway during leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1854. [Google Scholar] [CrossRef] [Green Version]
- Wada, S.; Ishida, H.; Izumi, M.; Yoshimoto, K.; Ohsumi, Y.; Mae, T.; Makino, A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2008, 149, 885–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, P.O.; Woo, H.R.; Nam, H.G. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003, 8, 272–278. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef] [Green Version]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [Green Version]
- Penfold, C.A.; Buchanan-Wollaston, V. Modelling transcriptional networks in leaf senescence. J. Exp. Bot. 2014, 65, 3859–3873. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, H.J.; Vu, Q.T.; Jung, S.; McClung, C.R.; Hong, S. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 8448–8453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 1511–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef] [Green Version]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Chai, J.; Liu, J.; Zhou, J.; Xing, D. Mitogen-activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. J. Exp. Bot. 2014, 65, 6513–6528. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.; Wang, R.; Chen, Y.; Shu, S.; Huang, R.; Zhang, D.; Li, J.; Xiao, S.; Yao, N.; et al. The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 2017, 173, 2294–2307. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.G.; Sun, C.H.; Zhang, Q.Y.; Gu, K.D.; Hao, Y.J. The basic helix-loop-helix transcription factor MdbHLH3 modulates leaf senescence. in apple via the regulation of dehydratase-enolase-phosphatase complex 1. Hortic. Res. 2020, 7, 50. [Google Scholar] [CrossRef] [Green Version]
- Budziszewski, G.J.; Lewis, S.P.; Glover, L.W.; Reineke, J.; Jones, G.; Ziemnik, L.S.; Levin, J.Z. Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 2001, 159, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Ecker, J.R. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, Y.J.; Ji, M.; Lee, A.; Killens, R.; Grunden, A.M.; Boss, W.F. Expression of Pyrococcus furiosus superoxide reductase in Arabidopsis enhances heat tolerance. Plant Physiol. 2009, 151, 893–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronsson, H.; Boij, P.; Patel, R.; Wardle, A.; Töpel, M.; Jarvis, P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007, 52, 53–68. [Google Scholar] [CrossRef]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef] [Green Version]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Mohanty, S.; Tripathy, B.C. Role of temperature stress on chloroplast biogenesis and protein import in pea. Plant Physiol. 2009, 150, 1050–1061. [Google Scholar] [CrossRef]
- Li, S.; Fu, Q.; Huang, W.; Yu, D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009, 28, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.M.; Mattoon, E.M.; Zhang, N.; Becker, E.; McHargue, W.; Yang, J.; Patel, D.; Dautermann, O.; McAdam, S.A.M.; Tarin, T.; et al. High light and temperature reduce photosynthetic efficiency through different mechanisms in the C4 model Setaria viridis. Commun. Biol. 2021, 4, 1092–1111. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, H.; Combe, J.; Patel, R.; Jarvis, P. In vivo assessment of the significance of phosphorylation of the Arabidopsis chloroplast protein import receptor, atToc33. FEBS Lett. 2006, 580, 649–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, R.; Hirashima, M.; Satoh, S.; Tanaka, A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: Inhibition of the pheophorbide an oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 2003, 44, 1266–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubry, S.; Fankhauser, N.; Ovinnikov, S.; Pružinská, A.; Stirnemann, M.; Zienkiewicz, K.; Herrfurth, C.; Feussner, I.; Hörtensteiner, S. Pheophorbide a May Regulate Jasmonate Signaling during Dark-Induced Senescence. Plant Physiol. 2019, 182, 776–791. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.; Close, P.S.; Briggs, S.P.; Johal, G.S. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 1997, 89, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.; Wardzala, E.; Yang, M.; Reinbothe, S.; Haller, S.; Pauli, F. A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol. Biol. 2004, 54, 39–54. [Google Scholar] [CrossRef]
- Liebsch, D.; Keech, O. Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytol. 2016, 212, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.R.; Suttangkakul, A.; Vierstra, R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008, 178, 1339–1353. [Google Scholar] [CrossRef] [Green Version]
- Weaver, L.M.; Amasino, R.M. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 2001, 127, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Lou, Y.; Shi, H.; Chen, L.; Cao, L. Transcriptomic analysis of dark-induced senescence in Bermudagrass (Cynodon dactylon). Plants 2019, 8, 614. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 2019, 222, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, 1154–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Zentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–177. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 2014, 5, 650. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef] [Green Version]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [Green Version]
- Murre, C.; Bain, G.; van Dijk, M.A.; Engel, I.; Furnari, B.A.; Massari, M.E.; Matthews, J.R.; Quong, M.W.; Rivera, R.R.; Stuiver, M.H. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta 1994, 1218, 129–135. [Google Scholar] [CrossRef]
- Quong, M.W.; Massari, M.E.; Zwart, R.; Murre, C. A new transcriptional-activation motif restricted to a class of helix–loop–helix proteins are functionally conserved in both yeast and mammalian cells. Mol. Cell Biol. 1993, 13, 792–800. [Google Scholar] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]







| Target Genes | Primer Name | Oligo Sequence (5′ to 3′) | Purpose |
|---|---|---|---|
| PED1 | PED1-RT-F | GCAATCATGGGTATCGGTCCAG | Semi qRT-PCR |
| PED1-RT-R | CTAGCGAGCGTCCTTGGACAAA | Semi qRT-PCR | |
| SEN1 | SEN1-RT-F | ATCACGAATTGGAAACTGG | Semi qRT-PCR |
| SEN1-RT-R | CTTTCCTCCATCGGAAG | Semi qRT-PCR | |
| BCB | BCB-RT-F | GAGGCATGATGTGGCAGTTGTAT | Semi qRT-PCR |
| BCB-RT-R | TCAAAAGAGAGCAACAACAGCAG | Semi qRT-PCR | |
| RBCS2B | RBCS2B-RT-F | ACCTTCTCCGCAACAAGTGG | Semi qRT-PCR |
| RBCS2B-RT-R | GAAGCTTGGTGGCTTGTAGG | Semi qRT-PCR | |
| Actin2 | ACT2-RT-F | ACATTGCAAAGAGTTTCAAGGT | Semi qRT-PCR |
| ACT2-RT-R | CTAAGCTCTCAAGATCAAAGG | Semi qRT-PCR | |
| AtbHLH19 | pGADT7-F | TAATACGACTCACTATAGGGC | Colony PCR |
| (AtbHLH41, AtbHLH92, AtbHLH133) | pGADT7-R | AGATGGTGCACGATGCACAG | Colony PCR |
| ANAC003p | pHis2.1-F | CTATAGGGCGAATTCGAG | Colony PCR |
| (FL, △1, △2) | pHis2.1-R | GATAATGCCAGGAATTACTAG | Colony PCR |
| Locus ID | GO Term | Annotation | Expression | Fold Change (tic55-II/WT) |
|---|---|---|---|---|
| NM_100103 | GO:0010228~vegetative to reproductive phase transition of meristem GO:0003700~transcription factor activity GO:0016926~ protein desumoylation GO:0007275~multicellular organism development GO:0005634~ nucleus GO:0050665~hydrogen peroxide biosynthetic process | A. thaliana NAC domain containing protein 3 mRNA (ANAC003) | Down | −2.237 |
| NM_102615 | GO:0010228~ vegetative to reproductive phase transition of meristem GO:0003700~ transcription factor activity GO:2000652~ regulation of secondary cell wall biogenesis GO:0016926~ protein desumoylation GO:0007275~ multicellular organism development GO:0005634~ nucleus GO:0050665~ hydrogen peroxide biosynthetic process GO:0045893~ positive regulation of transcription, DNA-templated | A. thaliana NAC domain containing protein 10 mRNA (ANAC010) | Down | −5.360 |
| NM_129861 | GO:1900056~ negative regulation of leaf senescence GO:0034976~ response to endoplasmic reticulum stress GO:0003700~ transcription factor activity GO:0009718~ anthocyanin- containing compound biosynthetic process GO:0005634~ nucleus GO:0007275~ multicellular organism development GO:0042538~ hyperosmotic salinity response GO:0010120~ camalexin biosynthetic process GO:0010150~ leaf senescence GO:0009627~systemic acquired resistance GO:0005992~ trehalose biosynthetic process GO:0006561~ proline biosynthetic process GO:0009723~ response to ethylene | A. thaliana NAC domain containing protein 42 mRNA (ANAC042) | Down | −2.244 |
| NM_119067 | GO:0003700~transcription factor activity GO:0007275~multicellular organism development GO:0005634~ nucleus | A. thaliana NAC domain containing protein 75 mRNA (ANAC075) | Down | −3.109 |
| NM_118328 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:0030528~ transcription regulator activity GO:0043565~ sequence-specific DNA binding | A. thaliana WRKY DNA-binding protein 31 mRNA (AtWRKY31) | Down | −3.310 |
| NM_001084564 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:0030528~ transcription regulator activity GO:0043565~ sequence-specific DNA binding | A. thaliana WRKY transcription factor 55 mRNA (AtWRKY55) | Down | −2.839 |
| NM_121517 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:0030528~ transcription regulator activity GO:0043565~ sequence-specific DNA binding | A. thaliana putative WRKY transcription factor 72 mRNA (AtWRKY72) | Down | −7.990 |
| NM_121311 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:0030528~ transcription regulator activity GO:0043565~ sequence-specific DNA binding | A. thaliana putative WRKY transcription factor 75 mRNA (AtWRKY75) | Down | −2.627 |
| NM_127841 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:005634~ nucleus GO:006355~ regulation of transcription, DNA-templated GO:0010200~ response to chitin | A. thaliana transcription factor bHLH19 mRNA (AtbHLH19) | Down | −3.046 |
| NM_125078 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:005634~ nucleus GO:006355~ regulation of transcription, DNA-templated | A. thaliana transcription factor bHLH41 mRNA (AtbHLH41) | Down | −3.677 |
| NM_123731 | GO:0003677~ DNA binding GO:0003700~ transcription factor activity GO:005634~ nucleus | A. thaliana transcription factor bHLH92 mRNA (AtbHLH92) | Down | −4.638 |
| NM_127568 | GO:005634~ Nucleus | A. thaliana transcription factor bHLH133 mRNA (AtbHLH133) | Down | −2.626 |
| NAC Genes | AtWRKY Binding Sequences (Nucleotides No.) 1 |
|---|---|
| ANAC003 | TGACT (−207~−203); TGACT (−399~−403); TTGACA (−755~−750) TTGACC (−876~-881); TTGACG (-949~-954) |
| ANAC010 | TGACC (−32~−36); TTGACG (−222~−217); TGACT (−294~−298) TTGACA (−927~−932) |
| ANAC042 | TGACT (−39~−35); TTGACC (−130~-125); TTTGACT (−143~−149) TGACT (−789~−785) |
| ANAC075 | TGACC (−623~−627) |
| NAC Genes | AtbHLH Binding Sequences (Nucleotides No.) 1 |
|---|---|
| ANAC003 | AGACGTAT (−113~−119); ATACTTGT (−216~−222); ATACGTGG (−279~−286); CAAGTG (−305~−300); ACACGTAA (−524~−531); CAAATG (−599~−594); ATACTTTT (−717~−710); GCAAGTTC (−780~−773); CAAATGCATATG (−785~−796); CAAATG (−824~−829); CAATTG (−833~−838); ACCAGT (−855~−860); ATTCGTGG (−873~−866); ATATGAGT (−875~−882); CATATG (−888~−893); TTACGTTT (−972~−965) |
| ANAC010 | CAAGTG (−28~−33); CAAGTG (−124~−129); ATACGTTT (−244~−251); CAACTG (−343~−338); CATATG (−613~−608); ATACATGA (−748~-741); ACCAAGTTGGT (−839~−829); CACGAG (−882~−877) |
| ANAC042 | TGACT (−39~−35); TTGACC (−130~−125); TTTGACT (−143~−149) TGACT (−789~−785) |
| ANAC075 | TGACC (−623~−627) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, C.-Y.; Chou, M.-L.; Wei, W.-C.; Chung, Y.-C.; Loo, X.-Y.; Lin, L.-F. Chloroplast Protein Tic55 Involved in Dark-Induced Senescence through AtbHLH/AtWRKY-ANAC003 Controlling Pathway of Arabidopsis thaliana. Genes 2022, 13, 308. https://doi.org/10.3390/genes13020308
Hsu C-Y, Chou M-L, Wei W-C, Chung Y-C, Loo X-Y, Lin L-F. Chloroplast Protein Tic55 Involved in Dark-Induced Senescence through AtbHLH/AtWRKY-ANAC003 Controlling Pathway of Arabidopsis thaliana. Genes. 2022; 13(2):308. https://doi.org/10.3390/genes13020308
Chicago/Turabian StyleHsu, Chou-Yu, Ming-Lun Chou, Wan-Chen Wei, Yo-Chia Chung, Xin-Yue Loo, and Lee-Fong Lin. 2022. "Chloroplast Protein Tic55 Involved in Dark-Induced Senescence through AtbHLH/AtWRKY-ANAC003 Controlling Pathway of Arabidopsis thaliana" Genes 13, no. 2: 308. https://doi.org/10.3390/genes13020308






