PAs Regulate Early Somatic Embryo Development by Changing the Gene Expression Level and the Hormonal Balance in Dimocarpus longan Lour.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of Genes Involved in PA Biosynthesis and Metabolism in Longan
2.3. Collinearity Analysis of the Genes Involved in PA Biosynthesis and Metabolism
2.4. Phylogenetic Analysis of the Gene Families Involved in PA Biosynthesis and Metabolism
2.5. The Analysis of Fpkm Expression Profile of Genes Involved in PA Biosynthesis and Metabolism
2.6. Determination of Endogenous Hormone Levels and H2O2 Content in Longan Treated with Exoge-Nous ACC, D-arg and PAs
2.7. RNA Extraction and Real-Time Fluorescence Quantitative Polymerase Chain Reaction (qRT-Pcr)
3. Results
3.1. Identification of Genes Involved in PA Biosynthesis and Metabolism
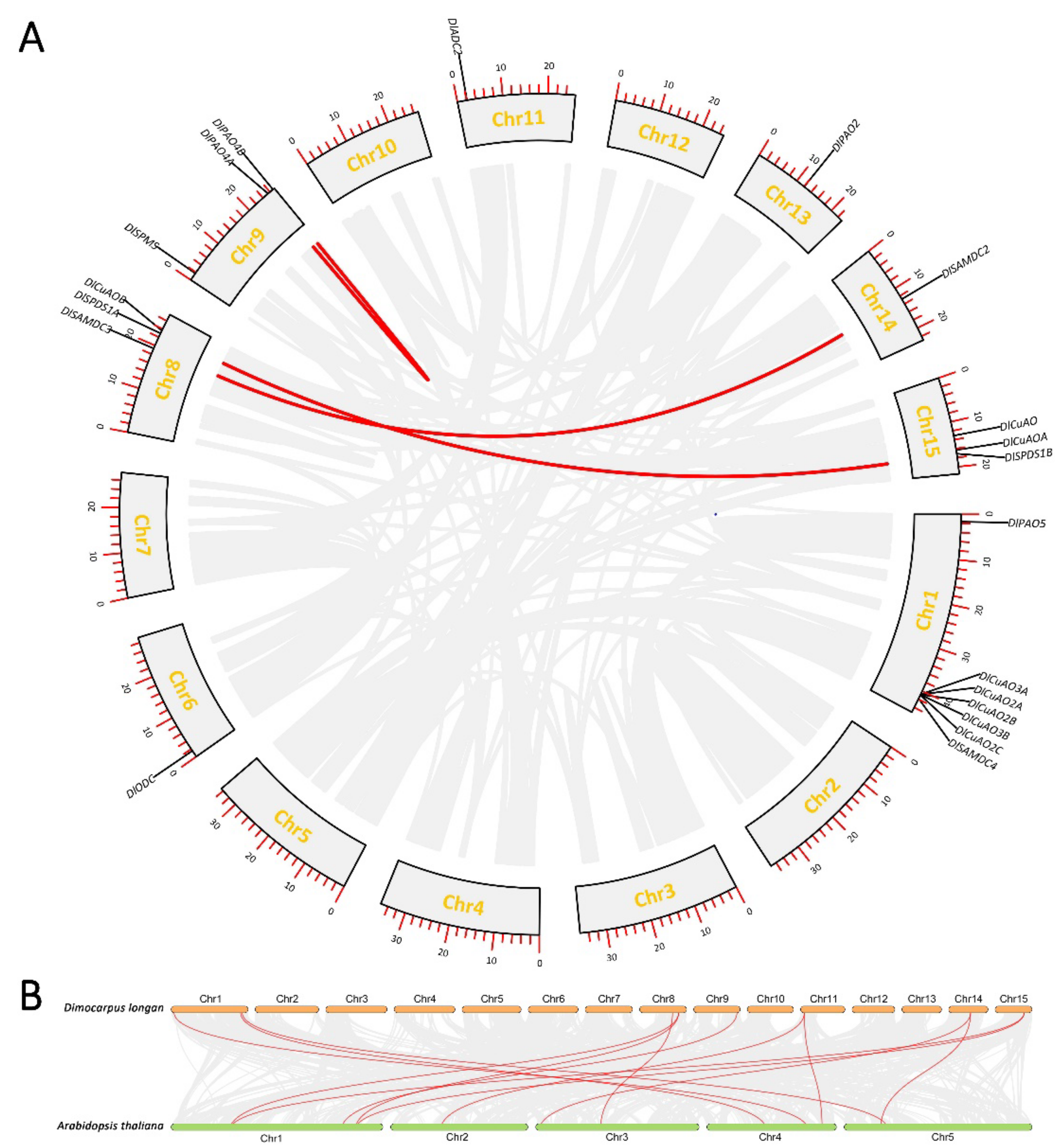
3.2. Chromosomal Localization and Collinearity Analysis of Genes Involved in PA Biosynthesis and Metabolism
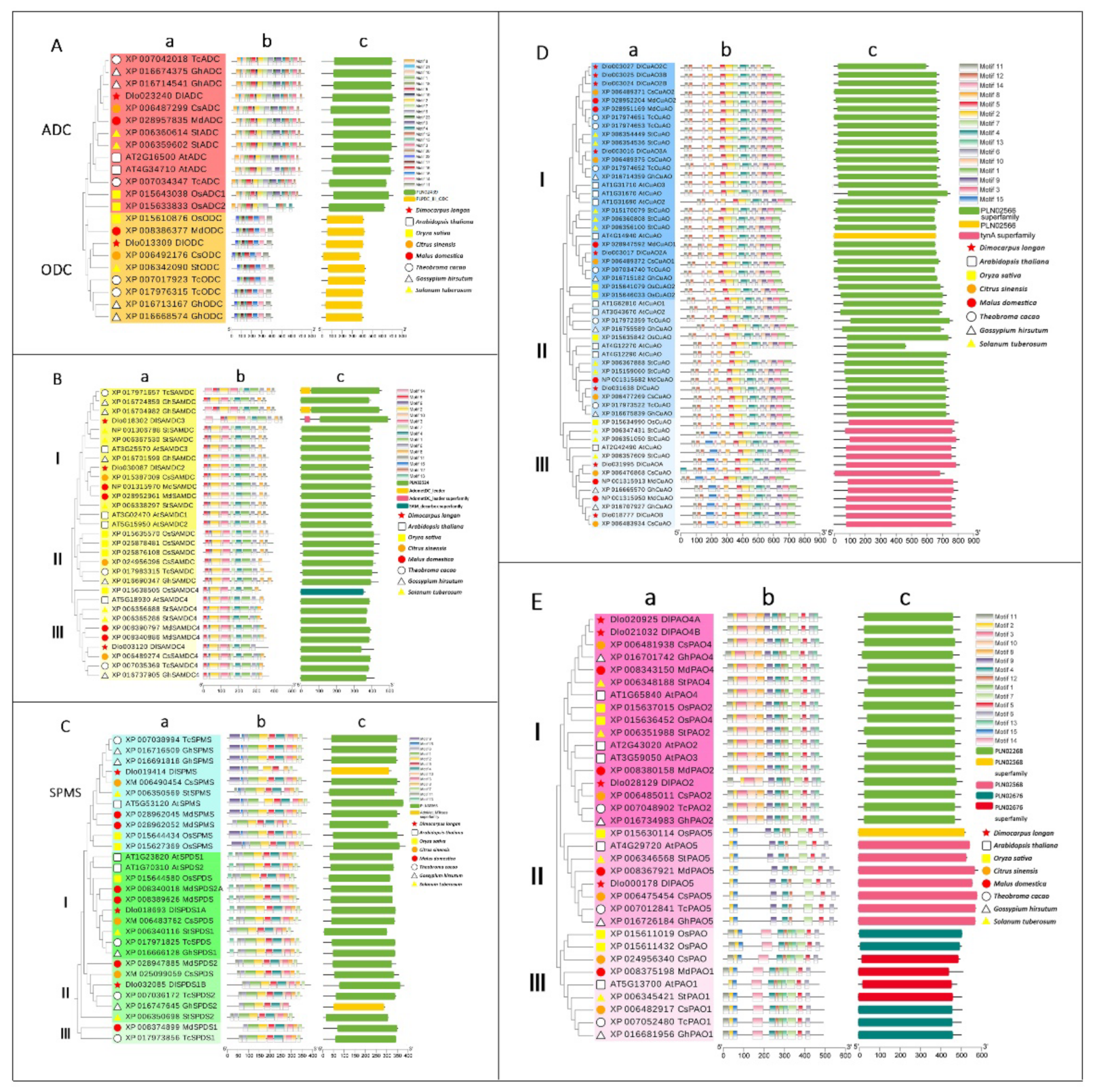
3.3. Phylogenetic Trees, Conserved Motifs and Conserved Domain Analysis of Gene Families Involved in PA Biosynthesis and Metabolism
3.4. Analysis of Structure and Cis-Acting Elements of Genes Involved in PA Biosynthesis and Metabolism in Longan
3.5. Expression Analysis of PA Biosynthesis and Metabolism Genes during Early SE in Longan Genome
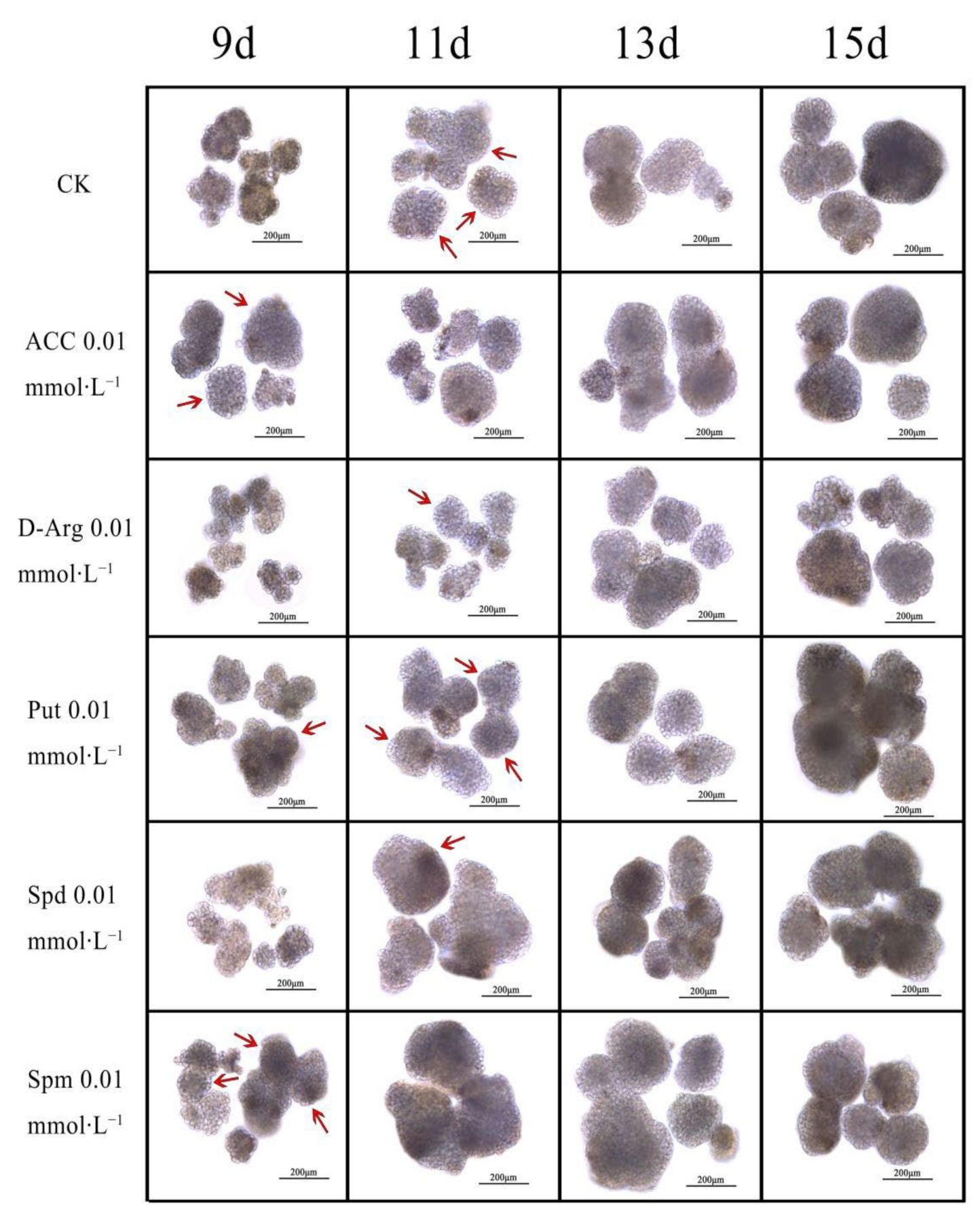
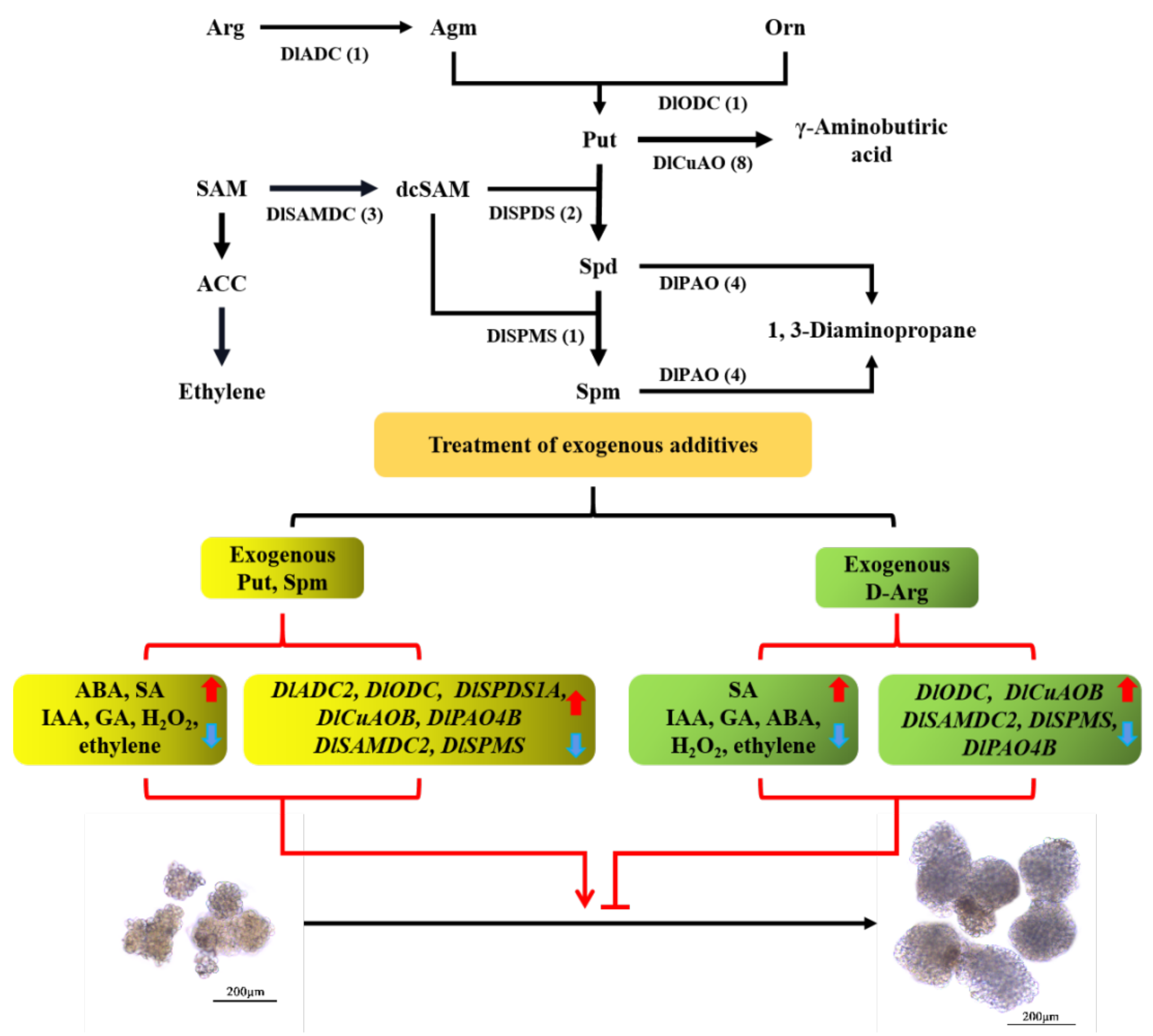
3.6. Effects of Exogenous ACC, D-arg and PAs on the Conversion of EC into GE in Longan
3.7. Effects of Exogenous ACC, D-arg and PAs on Endogenous Hormone Level and H2O2 Content in Longan
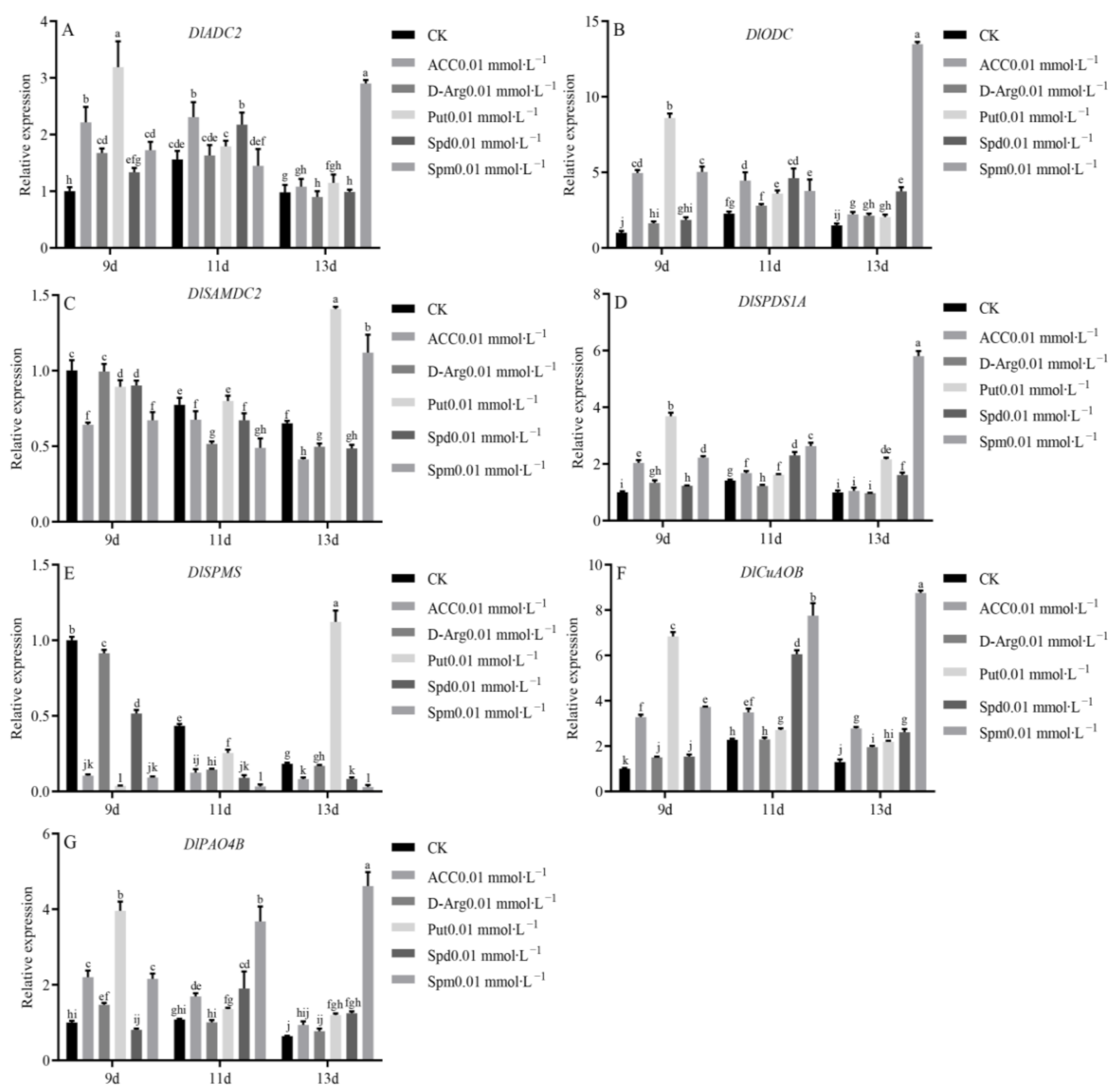
3.8. Expression Analysis of PA Biosynthesis and Metabolism Family Members under Exogenous ACC, D-arg and PAs Treatment
4. Discussion
4.1. Structural and Evolutionary Analysis of Members of PA Biosynthesis and Metabolism Family
4.2. Exogenous ACC, D-arg and PAs Regulate the Morphogenesis of Longan GE by Changing the Level of Endogenous Hormones
4.3. Exogenous ACC, D-arg and PAs Were Involved in Regulating the Expression of PA Biosynthesis and Metabolic Family Members in Longan
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kusano, T.; Yamaguchi, K.; Berberich, T.; Takahashi, Y. Advances in polyamine research in 2007. J. Plant Res. 2007, 120, 345–350. [Google Scholar] [CrossRef]
- Vuosku, J.; Suorsa, M.; Ruottinen, M.; Sutela, S.; Muilu-Makela, R.; Julkunen-Tiitto, R.; Sarjala, T.; Neubauer, P.; Haggman, H. Polyamine metabolism during exponential growth transition in Scots pine embryogenic cell culture. Tree Physiol. 2012, 32, 1274–1287. [Google Scholar] [CrossRef] [Green Version]
- Alcazar, R.; Altabella, T.; Marco, F.; Bortolotti, C.; Reymond, M.; Koncz, C.; Carrasco, P.; Tiburcio, A.F. Polyamines: Molecules with regulatory functions in plant abiotic stress tolerance. Planta 2010, 231, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Planas, J.; Saxena, T.; Zarza, X.; Bortolotti, C.; Cuevas, J.; Bitrián, M.; Tiburcio, A.F.; Altabella, T. Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants over-expressing the homologous Arginine decarboxylase 2 gene. Plant Physiol. Bioch. 2010, 48, 547–552. [Google Scholar] [CrossRef]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Francis, R. Polyamines and apoptosis. J. Cell. Mol. Med. 2005, 9, 623–642. [Google Scholar]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines regulate strawberry fruit ripening by abscisic acid, auxin, and ethylene. Plant Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Zhou, W.; Liang, X.; Zhou, K.; Lin, X. Increased bound putrescine accumulation contributes to the maintenance of antioxidant enzymes and higher aluminum tolerance in wheat. Environ. Pollut. 2019, 252, 941–949. [Google Scholar] [CrossRef]
- Liu, J.; Nada, K.; Honda, C.; Kitashiba, H.; Wen, X.; Pang, X.; Moriguchi, T. Polyamine biosynthesis of apple callus under salt stress: Importance of the arginine decarboxylase pathway in stress response. J. Exp. Bot. 2006, 57, 2589–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagor, G.H.M.; Simm, S.; Kim, D.W.; Niitsu, M.; Kusano, T.; Berberich, T. Effect of thermospermine on expression profiling of different gene using massive analysis of cDNA ends (MACE) and vascular maintenance in Arabidopsis. Physiol. Mol. Biol. Plants 2021, 27, 577–586. [Google Scholar] [CrossRef]
- Gong, X.; Dou, F.; Cheng, X.; Zhou, J.; Zou, Y.; Ma, F. Genome-wide identification of genes involved in polyamine biosynthesis and the role of exogenous polyamines in Malus hupehensis Rehd. under alkaline stress. Gene 2018, 669, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, X.; Liu, S.; Tan, B.; Cheng, J.; Ye, X.; Li, J.; Feng, J. Polyamine oxidase (PAO)–mediated polyamine catabolism plays potential roles in peach (Prunus persica L.) fruit development and ripening. Tree Genet. Genomes 2021, 17, 1–15. [Google Scholar] [CrossRef]
- Torrigiani, P.; Bregoli, A.M.; Ziosi, V.; Costa, G. Molecular and biochemical aspects underlying polyamine modulation of fruit development and ripening. Stewart Postharvest Rev. 2008, 4, 1–12. [Google Scholar] [CrossRef]
- Pál, M.; Szalai, G.; Gondor, O.K.; Janda, T. Unfinished story of polyamines: Role of conjugation, transport and light-related regulation in the polyamine metabolism in plants. Plant Sci. 2021, 308, 110923. [Google Scholar] [CrossRef] [PubMed]
- Fraudentali, I.; Rodrigues-Pousada, R.A.; Tavladoraki, P.; Angelini, R.; Cona, A. Leaf-wounding long-distance signaling targets AtCuAOβ leading to root phenotypic plasticity. Plants 2020, 9, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Zou, Y.; Wu, Q.; Kuča, K. Arbuscular mycorrhizas modulate root polyamine metabolism to enhance drought tolerance of trifoliate orange. Environ. Exp. Bot. 2020, 171, 103926. [Google Scholar] [CrossRef]
- Yuan, H.; Yue, P.; Bu, H.; Han, D.; Wang, A. Genome-wide analysis of ACO and ACS genes in pear (Pyrus ussuriensis). In vitro Cell. Dev. Biol.-Plant 2020, 56, 193–199. [Google Scholar] [CrossRef]
- Shu, S.; Chen, L.; Lu, W.; Sun, J.; Guo, S.; Yuan, Y.; Li, J. Effects of exogenous spermidine on photosynthetic capacity and expression of Calvin cycle genes in salt-stressed cucumber seedlings. J. Plant Res. 2014, 127, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhang, J.; Hu, J.; Wang, W.; Wu, H.; Zhang, Q.; Liu, J. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015, 38, 2248–2262. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Peng, D.; Peng, Y.; Zhang, X.; Ma, X.; Huang, L.; Yan, Y. The inhibition of polyamine biosynthesis weakens the drought tolerance in white clover (Trifolium repens) associated with the alteration of extensive proteins. Protoplasma 2018, 255, 803–817. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Lee, I. Ameliorative effects of spermine against osmotic stress through antioxidants and abscisic acid changes in soybean pods and seeds. Acta Physiol. Plant. 2013, 35, 263–269. [Google Scholar] [CrossRef]
- Galston, A.W. Polyamines as modulators of plant development. BioScience 2013, 33, 382–388. [Google Scholar] [CrossRef]
- Niemi, K.; Sarjala, T.; Chen, X.; Häggman, H. Spermidine and methylglyoxal bis (guanylhydrazone) affect maturation and endogenous polyamine content of Scots pine embryogenic cultures. J. Plant Physiol. 2002, 159, 1155–1158. [Google Scholar] [CrossRef]
- Kumar, V.; Giridhar, P.; Chandrashekar, A.; Ravishankar, G.A. Polyamines influence morphogenesis and caffeine biosynthesis in in vitro cultures of Coffea canephora P. ex Fr. Acta Physiol. Plant. 2008, 30, 217–223. [Google Scholar] [CrossRef]
- Mauri, P.V.; Manzanera, J.A. Somatic embryogenesis of holm oak (Quercus ilex L.): Ethylene production and polyamine content. Acta Physiol. Plant. 2011, 33, 717–723. [Google Scholar] [CrossRef]
- Cheng, W.; Wang, F.; Cheng, X.; Zhu, Q.; Sun, Y.; Zhu, H.; Sun, J. Polyamine and its metabolite H2O2 Play a key role in the conversion of embryogenic callus into somatic embryos in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 2015, 6, 1063. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.L.; Min, J.M.; Lai, R.L.; Wu, Z.Y.; Chen, Y.K.; Yu, L.L.; Cheng, C.Z.; Jin, Y.C.; Tian, Q.L.; Liu, Q.F.; et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. GigaScience 2017, 6, gix023. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.Y.; Chen, W. Relationship between embryonic development and changes of endogenous polyamines in longan (Dimocarpus Longan Lour.) ovules. Chin. J. Appl. Environ. Biol. 2005, 11, 286–288. [Google Scholar] [CrossRef]
- Chen, C.L.; Lai, Z.X. The changes of the contents of endogenous polyamines in the process of somatic embryogenesis from embryogenic callus in longan. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2006, 35, 381–383. [Google Scholar] [CrossRef]
- Li, R. Studies on Proteomics during Friable Embryogenic Callus Development in Dimocarpus longan Lour.; Fujian Agriculture and Forestry University: Fuzhou, China, 2011. [Google Scholar]
- Chen, J.Z.; Liu, Y.C.; Ye, Z.X.; Hu, Y.L.; Feng, Q.R. Changes in endogenous polyamine contents during fruit development of longan (Dimocarpus Longan Lour.). Chin. J. Trop. Crops 2006, 27, 18–23. [Google Scholar] [CrossRef]
- Lai, Z.X.; Pan, L.Z.; Chen, Z.G. Establishment and maintenance of longan embryogenic cell lines. J. Fujian Agric. University. 1997, 02, 33–40. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic. Acids Res. 2019, 47, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, X.; Shen, X.; Chen, R.; Zhu, C.; Zhang, Z.; Chen, Y.; Lin, W.; Xu, X.; Lin, Y.; et al. Genome-wide identification and characterization of DEAD-box helicase family associated with early somatic embryogenesis in Dimocarpus longan Lour. J. Plant Physiol. 2021, 258–259, 153364. [Google Scholar] [CrossRef] [PubMed]
- Mirza, H.; Alhaithloul, H.A.S.; Parvin, K.; Bhuyan, M.H.M.B.; Tanveer, M.; Mohsin, S.M.; Nahar, K.; Soliman, M.H.; Mahmud, J.A.; Fujita, M. Polyamine action under metal/metalloid stress: Regulation of biosynthesis, metabolism, and molecular interactions. Int. J. Mol. Sci. 2019, 20, 3215. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Hou, J.; Gao, W.; Tong, X.; Li, M.; Chu, X.; Chen, G. Exogenous spermidine alleviates the adverse effects of aluminum toxicity on photosystem II through improved antioxidant system and endogenous polyamine contents. Ecotox. Environ. Safe. 2021, 207, 111265. [Google Scholar] [CrossRef]
- Li, Y.C.; Ma, Y.Y.; Zhang, T.T.; Bi, Y.; Wang, Y.; Prusky, D. Exogenous polyamines enhance resistance to Alternaria alternata by modulating redox homeostasis in apricot fruit. Food Chem. 2019, 301, 125303. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Zhang, X.; Yan, J.; Fan, L.; Rong, C.; Mo, C.; Zhang, M. Effect of exogenous spermidine on floral induction, endogenous polyamine and hormone production, and expression of related genes in “Fuji” apple (Malus domestica Borkh.). Sci. Rep.-UK 2019, 9, 12777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, X.; Zhan, J.; Pan, J.; He, M.; Li, B.; Wang, J.; Ma, H.; Wang, Y.; Liu, S. The rational discovery of multipurpose inhibitors of the ornithine decarboxylase. FASEB J. 2020, 34, 10907–12921. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.B.; Emory, K.K.; Piatak, R.M.; Malmberg, R.L. Arginine decarboxylase (polyamine synthesis) mutants of Arabidopsis thaliana exhibit altered root growth. Plant J. Cell Mol. Biol. 1998, 13, 231–239. [Google Scholar] [CrossRef] [PubMed]
- He, L.M.U.T.; Nada, K.; Kasukabe, Y.; Tachibana, S. Enhanced susceptibility of photosynthesis to low-temperature photoinhibition due to interruption of chill-induced increase of S-adenosylmethionine decarboxylase activity in leaves of spinach (Spinacia oleracea L.). Plant Cell Physiol. 2002, 43, 196–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuosku, J.; Karppinen, K.; Muilu-Mäkelä, R.; Kusano, T.; Sagor, G.H.M.; Avia, K.; Alakärppä, E.; Kestilä, J.; Suokas, M.; Nickolov, K.; et al. Scots pine aminopropyltransferases shed new light on evolution of the polyamine biosynthesis pathway in seed plants. Ann. Bot. 2018, 121, 1243–1256. [Google Scholar] [CrossRef] [Green Version]
- Fraudentali, I.; Ghuge, S.A.; Carucci, A.; Tavladoraki, P.; Angelini, R.; Cona, A.; Rodrigues-Pousada, R.A. The copper amine oxidase AtCuAOδ participates in abscisic acid-induced stomatal closure in Arabidopsis. Plants 2019, 8, 183. [Google Scholar] [CrossRef] [Green Version]
- Qu, Y.; Liu, X.; Zhang, X.; Tang, Y.; Hu, Y.; Chen, S.; Xiang, L.; Zhang, Q. Transcriptional regulation of Arabidopsis copper amine oxidase ζ (CuAOζ) in indole-3-butyric acid-induced lateral root development. Plant Growth Regul. 2019, 89, 287–297. [Google Scholar] [CrossRef]
- Pal, M.; Csavas, G.; Szalai, G.; Olah, T.; Khalil, R.; Yordanova, R.; Gell, G.; Birinyi, Z.; Nemeth, E.; Janda, T. Polyamines may influence phytochelatin synthesis during Cd stress in rice. J. Hazard. Mater. 2017, 340, 272–280. [Google Scholar] [CrossRef] [Green Version]
- Szalai, G.; Janda, K.; Darko, E.; Janda, T.; Peeva, V.; Pal, M. Comparative analysis of polyamine metabolism in wheat and maize plants. Plant Physiol. Biochem. 2017, 112, 239–250. [Google Scholar] [CrossRef] [Green Version]
- Tajti, J.; Janda, T.; Majláth, I.; Szalai, G.; Pál, M. Comparative study on the effects of putrescine and spermidine pre-treatment on cadmium stress in wheat. Ecotox. Environ. Safe. 2018, 148, 546–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doneva, D.; Pál, M.; Brankova, L.; Szalai, G.; Tajti, J.; Khalil, R.; Ivanovska, B.; Velikova, V.; Misheva, S.; Janda, T.; et al. The effects of putrescine pre-treatment on osmotic stress responses in drought-tolerant and drought-sensitive wheat seedlings. Physiol. Plant. 2021, 171, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Janse Van Rensburg, H.C.; Limami, A.M.; Van den Ende, W. Spermine and spermidine priming against Botrytis cinerea modulates ROS dynamics and metabolism in Arabidopsis. Biomolecules 2021, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zahng, Y.; Xiang, J.; Wu, H.; Chen, H.; Zhang, Y.; Zhu, D. Effects of chilling tolerance induced by spermidine pretreatment on antioxidative activity, endogenous hormones and ultrastructure of indica-japonica hybrid rice seedlings. J. Integr. Agr. 2016, 15, 295–308. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Lin, C.; He, F.; Li, Z.; Guan, Y.; Hu, Q.; Hu, J. Exogenous spermidine improves seed germination of sweet corn via involvement in phytohormone interactions, H2O2 and relevant gene expression. BMC Plant Biol. 2017, 17, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karami, O.; Saidi, A. The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol. Biol. Rep. 2010, 37, 2493–2507. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.Y.; Hu, L.S.; Yuan, D.J.; Xu, J.; Gao, W.H.; He, L.R.; Yang, X.Y.; Zhang, X.L. Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: Evidence for stress response functions in SE development. Plant Biotechnol. J. 2014, 12, 161–173. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Primer Sequences (5′ to 3′) | Produce Size (bp) | |
|---|---|---|---|
| DlADC2 | F: CCAGACCCACAACTACGAC | R: CAACCCGAAACGGAAAC | 115 |
| DLODC | F: GAGTGGCACCCGAAATG | R: CTTGAGCCGAGATGCTTG | 143 |
| DlSAMDC2 | F: CGTTACACCCGAAGATGG | R: GGAGCAAGCAAGAACCC | 106 |
| DlSPDS1A | F: CGCAGAGAGGGATGAATG | R: GCCGCCAATAACCAGAAC | 95 |
| DlSPMS | F: CGAATGCCACTCCACTG | R: CGTGTGCTTCTCCAGGC | 113 |
| DlCuAOB | F: CTTCTTCCCTCCATTCCAG | R: CACAACAAGTCTCGCACG | 97 |
| DlPAO4B | F: GTTGATACGCTCCCTTGG | R: GGAACTTTGTTGCCGTCC | 114 |
| DlFeSOD | F: CAGATGGTGAAGCCGTAGAG | R: TATGCCACCGATACAACAAAC | 106 |
| Gene Name | Second-Generation Genome ID | Third-Generation Genome ID | Annotated in Arabidopsis | Protein Length (aa) | MW (Da) | pI | II | AI | GRAVY | Signal Peptide | TMD | Phosphorylation Site | Subcellular Localization | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | Y | T | Total | |||||||||||||
| DlADC2 | Dlo_024362.1 | Dlo023240 | AtADC2 | 726 | 77813.0 | 5.23 | 41.67 | 90.32 | −0.043 | No | 0 | 43 | 11 | 10 | 64 | chloroplast |
| DlODC | Dlo_001547.1 | Dlo013309 | AtODC | 414 | 44583.8 | 5.94 | 40.39 | 87.22 | −0.007 | No | 0 | 16 | 4 | 11 | 31 | cytoplasm |
| DlSAMDC2 | Dlo_037470.1 | Dlo030087 | AtSAMDC2 | 362 | 40105.6 | 4.77 | 43.64 | 83.23 | −0.083 | No | 0 | 26 | 7 | 7 | 40 | chloroplast |
| DlSAMDC3 | Dlo_013246.1 | Dlo018302 | AtSAMDC3 | 451 | 50084.2 | 5.27 | 49.47 | 85.57 | −0.057 | No | 0 | 43 | 3 | 7 | 53 | chloroplast |
| DlSAMDC4 | Dlo_029928.1 | Dlo003120 | AtSAMDC4 | 368 | 40994.6 | 5.00 | 36.36 | 77.01 | −0.179 | No | 0 | 23 | 9 | 16 | 48 | cytoplasm |
| DlSPDS1A | Dlo_018546.3 | Dlo018693 | AtSPDS1 | 349 | 38036.2 | 4.90 | 42.10 | 82.87 | −0.136 | No | 0 | 15 | 3 | 10 | 28 | cytoplasm |
| DlSPDS1B | Dlo_012336.1 | Dlo032085 | AtSPDS1 | 390 | 43624.7 | 5.74 | 42.89 | 85.51 | −0.171 | No | 0 | 28 | 6 | 10 | 44 | chloroplast |
| DlSPMS | Dlo_032255.1 | Dlo019414 | AtSPMS | 328 | 35931.3 | 5.83 | 50.54 | 88.48 | −0.143 | No | 0 | 20 | 5 | 3 | 28 | cytoplasm |
| DlCuAO | Dlo_005522.1 | Dlo031638 | AtCuAO | 738 | 83634.3 | 6.14 | 40.72 | 80.41 | −0.348 | Yes | 1 | 31 | 9 | 25 | 65 | chloroplast |
| DlCuAOA | Dlo_019230.1 | Dlo031995 | AtCuAO | 806 | 89799.7 | 6.62 | 45.24 | 80.87 | −0.276 | No | 0 | 45 | 7 | 17 | 69 | chloroplast |
| DlCuAOB | Dlo_019230.1 | Dlo018777 | AtCuAO | 774 | 86787.0 | 6.69 | 45.31 | 78.18 | −0.372 | No | 0 | 29 | 4 | 19 | 52 | chloroplast |
| DlCuAO2A | Dlo_030965.1 | Dlo003017 | AtCuAO2 | 654 | 74021.1 | 6.75 | 36.00 | 84.02 | −0.247 | Yes | 0 | 29 | 9 | 25 | 63 | Plasma membrane |
| DlCuAO2B | Dlo_030970.1 | Dlo003024 | AtCuAO2 | 670 | 76074.0 | 6.15 | 34.77 | 83.31 | −0.287 | Yes | 1 | 25 | 7 | 22 | 54 | chloroplast |
| DlCuAO2C | Dlo_030974.1 | Dlo003027 | AtCuAO2 | 603 | 68096.8 | 5.97 | 34.38 | 86.29 | −0.254 | Yes | 0 | 25 | 4 | 24 | 53 | cytoplasm |
| DlCuAO3A | Dlo_030964.1 | Dlo003016 | AtCuAO3 | 672 | 76865.4 | 6.09 | 41.18 | 81.74 | −0.336 | No | 1 | 30 | 11 | 20 | 61 | cell wall |
| DlCuAO3B | Dlo_030971.1 | Dlo003025 | AtCuAO3 | 670 | 75847.6 | 6.49 | 30.81 | 83.90 | −0.288 | Yes | 0 | 25 | 8 | 27 | 60 | cell wall |
| DlPAO2 | Dlo_000974.2 | Dlo028129 | AtPAO2 | 498 | 55240.3 | 5.63 | 35.16 | 94.74 | −0.031 | No | 0 | 24 | 3 | 13 | 40 | chloroplast |
| DlPAO4A | Dlo_010639.1 | Dlo020925 | AtPAO4 | 486 | 53906.1 | 5.98 | 39.26 | 99.90 | −0.010 | No | 0 | 21 | 4 | 6 | 31 | chloroplast |
| DlPAO4B | Dlo_010639.1 | Dlo021032 | AtPAO4 | 486 | 53828.9 | 5.93 | 39.60 | 99.69 | −0.012 | No | 0 | 21 | 4 | 6 | 31 | chloroplast |
| DlPAO5 | Dlo_030297.1 | Dlo000178 | AtPAO5 | 546 | 60561.7 | 5.89 | 47.15 | 80.20 | −0.295 | Yes | 0 | 45 | 8 | 18 | 71 | cytoplasm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, C.; Zhou, X.; Zhang, S.; Zhang, X.; Liu, M.; Zhang, C.; Xu, X.; Xu, X.; Chen, X.; Chen, Y.; et al. PAs Regulate Early Somatic Embryo Development by Changing the Gene Expression Level and the Hormonal Balance in Dimocarpus longan Lour. Genes 2022, 13, 317. https://doi.org/10.3390/genes13020317
Lai C, Zhou X, Zhang S, Zhang X, Liu M, Zhang C, Xu X, Xu X, Chen X, Chen Y, et al. PAs Regulate Early Somatic Embryo Development by Changing the Gene Expression Level and the Hormonal Balance in Dimocarpus longan Lour. Genes. 2022; 13(2):317. https://doi.org/10.3390/genes13020317
Chicago/Turabian StyleLai, Chunwang, Xiaojuan Zhou, Shuting Zhang, Xueying Zhang, Mengyu Liu, Chunyu Zhang, Xiaoqiong Xu, Xiaoping Xu, Xiaohui Chen, Yan Chen, and et al. 2022. "PAs Regulate Early Somatic Embryo Development by Changing the Gene Expression Level and the Hormonal Balance in Dimocarpus longan Lour." Genes 13, no. 2: 317. https://doi.org/10.3390/genes13020317






