Methods to Improve Molecular Diagnosis in Genomic Cold Cases in Pediatric Neurology
Abstract
1. Introduction
2. Maximizing the Use of Information in Existing Sequence Data
2.1. Extended Analysis of Existing Data
2.2. Non-Mendelian Inheritance Models

2.3. Dual/Multiple Diagnoses
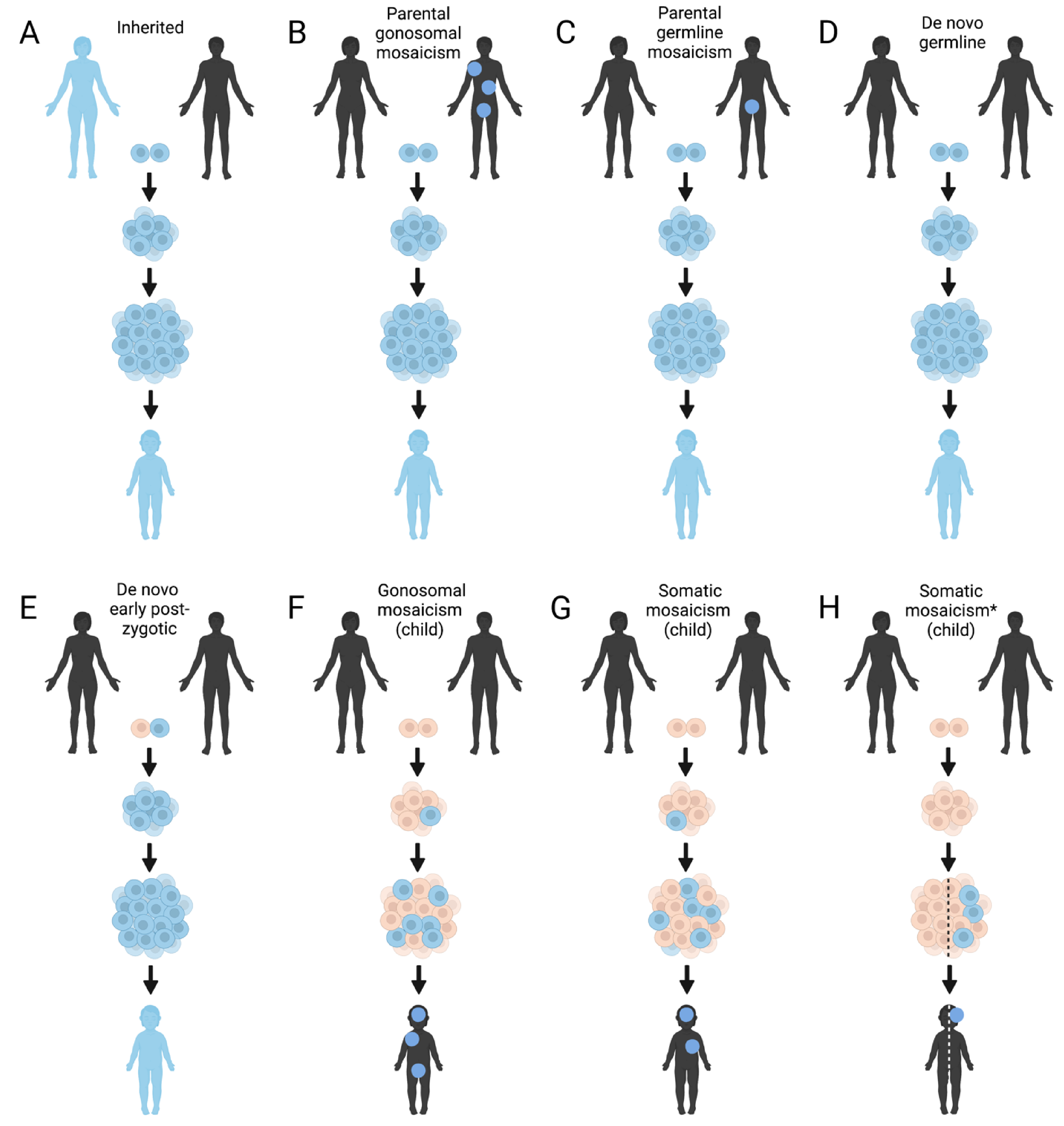
2.4. Mosaicism
3. Data Re-Analysis
3.1. Periodic Data Re-Analysis
3.2. Artificial Intelligence (AI) Applications
4. Integrating Omics to Understand Functional Effects and Improve Variant Prioritization
4.1. Transcriptomics
4.2. Epigenomics
4.3. Proteomics
4.4. Metabolomics
4.5. Public Recourses and Bioinformatic Predictions
5. Deep Phenotyping
6. Novel DNA Sequencing and Mapping Technologies
6.1. Long-Read Sequencing (LRS)
6.2. Artificial Long-Read Sequencing (Alrs)
6.3. Optical Genome Mapping (OGM)
6.4. Integrating Different Data Types
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilissen, C.; Hoischen, A.; Brunner, H.G.; Veltman, J.A. Unlocking Mendelian Disease Using Exome Sequencing. Genome Biol. 2011, 12, 228. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.B.; Marshall, J.L.; Tukiainen, T.; Lek, M.; Donkervoort, S.; Foley, A.R.; Bolduc, V.; Waddell, L.B.; Sandaradura, S.A.; O’Grady, G.L.; et al. Improving Genetic Diagnosis in Mendelian Disease with Transcriptome Sequencing. Sci. Transl. Med. 2017, 9, eaal5209. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Snyder, M.P. Integrative Omics for Health and Disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Stefanski, A.; Calle-López, Y.; Leu, C.; Pérez-Palma, E.; Pestana-Knight, E.; Lal, D. Clinical Sequencing Yield in Epilepsy, Autism Spectrum Disorder, and Intellectual Disability: A Systematic Review and Meta-Analysis. Epilepsia 2021, 62, 143–151. [Google Scholar] [CrossRef]
- Lionel, A.C.; Costain, G.; Monfared, N.; Walker, S.; Reuter, M.S.; Hosseini, S.M.; Thiruvahindrapuram, B.; Merico, D.; Jobling, R.; Nalpathamkalam, T.; et al. Improved Diagnostic Yield Compared with Targeted Gene Sequencing Panels Suggests a Role for Whole-Genome Sequencing as a First-Tier Genetic Test. Genet. Med. 2018, 20, 435–443. [Google Scholar] [CrossRef]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; van de Vorst, M.; van Bon, B.W.M.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A.; et al. Genome Sequencing Identifies Major Causes of Severe Intellectual Disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef]
- Bergant, G.; Maver, A.; Lovrecic, L.; Čuturilo, G.; Hodzic, A.; Peterlin, B. Comprehensive Use of Extended Exome Analysis Improves Diagnostic Yield in Rare Disease: A Retrospective Survey in 1,059 Cases. Genet. Med. 2018, 20, 303–312. [Google Scholar] [CrossRef]
- Pfundt, R.; del Rosario, M.; Vissers, L.E.L.M.; Kwint, M.P.; Janssen, I.M.; de Leeuw, N.; Yntema, H.G.; Nelen, M.R.; Lugtenberg, D.; Kamsteeg, E.-J.; et al. Detection of Clinically Relevant Copy-Number Variants by Exome Sequencing in a Large Cohort of Genetic Disorders. Genet. Med. 2017, 19, 667–675. [Google Scholar] [CrossRef]
- Suzuki, H.; Yamada, M.; Uehara, T.; Takenouchi, T.; Kosaki, K. Parallel Detection of Single Nucleotide Variants and Copy Number Variants with Exome Analysis: Validation in a Cohort of 700 Undiagnosed Patients. Am. J. Med. Genet. A 2020, 182, 2529–2532. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef]
- Krumm, N.; Sudmant, P.H.; Ko, A.; O’Roak, B.J.; Malig, M.; Coe, B.P.; NHLBI Exome Sequencing Project; Quinlan, A.R.; Nickerson, D.A.; Eichler, E.E. Copy Number Variation Detection and Genotyping from Exome Sequence Data. Genome Res. 2012, 22, 1525–1532. [Google Scholar] [CrossRef]
- Plagnol, V.; Curtis, J.; Epstein, M.; Mok, K.Y.; Stebbings, E.; Grigoriadou, S.; Wood, N.W.; Hambleton, S.; Burns, S.O.; Thrasher, A.J.; et al. A Robust Model for Read Count Data in Exome Sequencing Experiments and Implications for Copy Number Variant Calling. Bioinformatics 2012, 28, 2747–2754. [Google Scholar] [CrossRef]
- Fromer, M.; Moran, J.L.; Chambert, K.; Banks, E.; Bergen, S.E.; Ruderfer, D.M.; Handsaker, R.E.; McCarroll, S.A.; O’Donovan, M.C.; Owen, M.J.; et al. Discovery and Statistical Genotyping of Copy-Number Variation from Whole-Exome Sequencing Depth. Am. J. Hum. Genet. 2012, 91, 597–607. [Google Scholar] [CrossRef]
- Kokkonen, H.; Siren, A.; Määttä, T.; Kamila Kadlubowska, M.; Acharya, A.; Nouel-Saied, L.M.; Leal, S.M.; Järvelä, I.; Schrauwen, I. Identification of Microduplications at Xp21.2 and Xq13.1 in Neurodevelopmental Disorders. Mol. Genet. Genomic Med. 2021, 9, e1703. [Google Scholar] [CrossRef]
- Dong, X.; Liu, B.; Yang, L.; Wang, H.; Wu, B.; Liu, R.; Chen, H.; Chen, X.; Yu, S.; Chen, B.; et al. Clinical Exome Sequencing as the First-Tier Test for Diagnosing Developmental Disorders Covering Both CNV and SNV: A Chinese Cohort. J. Med. Genet. 2020, 57, 558–566. [Google Scholar] [CrossRef]
- Takumi, T.; Tamada, K. CNV Biology in Neurodevelopmental Disorders. Curr. Opin. Neurobiol. 2018, 48, 183–192. [Google Scholar] [CrossRef]
- Sun, Y.; Ye, X.; Fan, Y.; Wang, L.; Luo, X.; Liu, H.; Gao, X.; Gong, Z.; Wang, Y.; Qiu, W.; et al. High Detection Rate of Copy Number Variations Using Capture Sequencing Data: A Retrospective Study. Clin. Chem. 2020, 66, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Qi, M.; Wang, S.; Yuan, X. DINTD: Detection and Inference of Tandem Duplications from Short Sequencing Reads. Front Genet 2020, 11, 924. [Google Scholar] [CrossRef]
- Mohiyuddin, M.; Mu, J.C.; Li, J.; Bani Asadi, N.; Gerstein, M.B.; Abyzov, A.; Wong, W.H.; Lam, H.Y.K. MetaSV: An Accurate and Integrative Structural-Variant Caller for next Generation Sequencing. Bioinformatics 2015, 31, 2741–2744. [Google Scholar] [CrossRef]
- Thung, D.T.; de Ligt, J.; Vissers, L.E.; Steehouwer, M.; Kroon, M.; de Vries, P.; Slagboom, E.P.; Ye, K.; Veltman, J.A.; Hehir-Kwa, J.Y. Mobster: Accurate Detection of Mobile Element Insertions in next Generation Sequencing Data. Genome Biol. 2014, 15, 488. [Google Scholar] [CrossRef] [PubMed]
- English, A.C.; Salerno, W.J.; Hampton, O.A.; Gonzaga-Jauregui, C.; Ambreth, S.; Ritter, D.I.; Beck, C.R.; Davis, C.F.; Dahdouli, M.; Ma, S.; et al. Assessing Structural Variation in a Personal Genome-towards a Human Reference Diploid Genome. BMC Genomics 2015, 16, 286. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.S.; Urban, A.E.; Mills, R.E. Structural Variation in the Sequencing Era. Nat Rev Genet 2020, 21, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Boerwinkle, E.; Liu, X. In Silico Prediction of Splice-Altering Single Nucleotide Variants in the Human Genome. Nucleic Acids Res. 2014, 42, 13534–13544. [Google Scholar] [CrossRef]
- Lin, H.; Hargreaves, K.A.; Li, R.; Reiter, J.L.; Wang, Y.; Mort, M.; Cooper, D.N.; Zhou, Y.; Zhang, C.; Eadon, M.T.; et al. RegSNPs-Intron: A Computational Framework for Predicting Pathogenic Impact of Intronic Single Nucleotide Variants. Genome Biol. 2019, 20, 254. [Google Scholar] [CrossRef]
- Rentzsch, P.; Schubach, M.; Shendure, J.; Kircher, M. CADD-Splice—Improving Genome-Wide Variant Effect Prediction Using Deep Learning-Derived Splice Scores. Genome Med. 2021, 13, 31. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef]
- OMIM—Online Mendelian Inheritance in Man. Available online: https://www.omim.org/ (accessed on 18 June 2021).
- Renaux, A.; Papadimitriou, S.; Versbraegen, N.; Nachtegael, C.; Boutry, S.; Nowé, A.; Smits, G.; Lenaerts, T. ORVAL: A Novel Platform for the Prediction and Exploration of Disease-Causing Oligogenic Variant Combinations. Nucleic Acids Res. 2019, 47, W93–W98. [Google Scholar] [CrossRef]
- ORVAL—Oligogenic Resource for Variant AnaLysis. Available online: https://orval.ibsquare.be/ (accessed on 19 June 2021).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/ (accessed on 19 June 2021).
- Rahit, K.M.T.H.; Tarailo-Graovac, M. Genetic Modifiers and Rare Mendelian Disease. Genes 2020, 11, 239. [Google Scholar] [CrossRef]
- Yousaf, R.; Ahmed, Z.M.; Giese, A.P.; Morell, R.J.; Lagziel, A.; Dabdoub, A.; Wilcox, E.R.; Riazuddin, S.; Friedman, T.B.; Riazuddin, S. Modifier Variant of METTL13 Suppresses Human GAB1-Associated Profound Deafness. J. Clin. Investig. 2018, 128, 1509–1522. [Google Scholar] [CrossRef]
- Riazuddin, S.; Castelein, C.M.; Ahmed, Z.M.; Lalwani, A.K.; Mastroianni, M.A.; Naz, S.; Smith, T.N.; Liburd, N.A.; Friedman, T.B.; Griffith, A.J.; et al. Dominant Modifier DFNM1 Suppresses Recessive Deafness DFNB26. Nat. Genet. 2000, 26, 431–434. [Google Scholar] [CrossRef]
- Jacquemont, S.; Coe, B.P.; Hersch, M.; Duyzend, M.H.; Krumm, N.; Bergmann, S.; Beckmann, J.S.; Rosenfeld, J.A.; Eichler, E.E. A Higher Mutational Burden in Females Supports a “Female Protective Model” in Neurodevelopmental Disorders. Am. J. Hum. Genet 2014, 94, 415–425. [Google Scholar] [CrossRef]
- Marques, A.H.; O’Connor, T.G.; Roth, C.; Susser, E.; Bjørke-Monsen, A.-L. The Influence of Maternal Prenatal and Early Childhood Nutrition and Maternal Prenatal Stress on Offspring Immune System Development and Neurodevelopmental Disorders. Front. Neurosci. 2013, 7, 120. [Google Scholar] [CrossRef]
- Orten, D.J.; Fischer, S.M.; Sorensen, J.L.; Radhakrishna, U.; Cremers, C.W.R.J.; Marres, H.A.M.; Van Camp, G.; Welch, K.O.; Smith, R.J.H.; Kimberling, W.J. Branchio-Oto-Renal Syndrome (BOR): Novel Mutations in the EYA1 Gene, and a Review of the Mutational Genetics of BOR. Hum. Mutat. 2008, 29, 537–544. [Google Scholar] [CrossRef]
- Meng, H.; Xu, H.-Q.; Yu, L.; Lin, G.-W.; He, N.; Su, T.; Shi, Y.-W.; Li, B.; Wang, J.; Liu, X.-R.; et al. The SCN1A Mutation Database: Updating Information and Analysis of the Relationships among Genotype, Functional Alteration, and Phenotype. Hum. Mutat. 2015, 36, 573–580. [Google Scholar] [CrossRef]
- Goldberg-Stern, H.; Aharoni, S.; Afawi, Z.; Bennett, O.; Appenzeller, S.; Pendziwiat, M.; Kuhlenbäumer, G.; Basel-Vanagaite, L.; Shuper, A.; Korczyn, A.D.; et al. Broad Phenotypic Heterogeneity Due to a Novel SCN1A Mutation in a Family with Genetic Epilepsy with Febrile Seizures Plus. J. Child Neurol. 2014, 29, 221–226. [Google Scholar] [CrossRef]
- Del Gaudio, D.; Shinawi, M.; Astbury, C.; Tayeh, M.K.; Deak, K.L.; Raca, G. ACMG Laboratory Quality Assurance Committee Diagnostic Testing for Uniparental Disomy: A Points to Consider Statement from the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2020, 22, 1133–1141. [Google Scholar] [CrossRef]
- Nakka, P.; Pattillo Smith, S.; O’Donnell-Luria, A.H.; McManus, K.F.; 23andMe Research Team; Mountain, J.L.; Ramachandran, S.; Sathirapongsasuti, J.F. Characterization of Prevalence and Health Consequences of Uniparental Disomy in Four Million Individuals from the General Population. Am. J. Hum. Genet. 2019, 105, 921–932. [Google Scholar] [CrossRef]
- Benn, P. Uniparental Disomy: Origin, Frequency, and Clinical Significance. Prenat. Diagn. 2021, 41, 564–572. [Google Scholar] [CrossRef]
- Hoppman, N.; Rumilla, K.; Lauer, E.; Kearney, H.; Thorland, E. Patterns of Homozygosity in Patients with Uniparental Disomy: Detection Rate and Suggested Reporting Thresholds for SNP Microarrays. Genet. Med. 2018, 20, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, B.; Robberecht, W.; Van Den Bosch, L. RNA Toxicity in Non-Coding Repeat Expansion Disorders. EMBO J. 2020, 39, e101112. [Google Scholar] [CrossRef] [PubMed]
- Depienne, C.; Mandel, J.-L. 30 Years of Repeat Expansion Disorders: What Have We Learned and What Are the Remaining Challenges? Am. J. Hum. Genet. 2021, 108, 764–785. [Google Scholar] [CrossRef] [PubMed]
- Van Heyningen, V.; Yeyati, P.L. Mechanisms of Non-Mendelian Inheritance in Genetic Disease. Hum. Mol. Genet. 2004, 13, R225–R233. [Google Scholar] [CrossRef]
- Hoshina, N.; Johnson-Venkatesh, E.M.; Hoshina, M.; Umemori, H. Female-Specific Synaptic Dysfunction and Cognitive Impairment in a Mouse Model of PCDH19 Disorder. Science 2021, 372, eaaz3893. [Google Scholar] [CrossRef]
- Morissette, J.; Clépet, C.; Moisan, S.; Dubois, S.; Winstall, E.; Vermeeren, D.; Nguyen, T.D.; Polansky, J.R.; Côté, G.; Anctil, J.L.; et al. Homozygotes Carrying an Autosomal Dominant TIGR Mutation Do Not Manifest Glaucoma. Nat. Genet. 1998, 19, 319–321. [Google Scholar] [CrossRef]
- Balci, T.B.; Hartley, T.; Xi, Y.; Dyment, D.A.; Beaulieu, C.L.; Bernier, F.P.; Dupuis, L.; Horvath, G.A.; Mendoza-Londono, R.; Prasad, C.; et al. Debunking Occam’s Razor: Diagnosing Multiple Genetic Diseases in Families by Whole-Exome Sequencing. Clin. Genet. 2017, 92, 281–289. [Google Scholar] [CrossRef]
- Karaca, E.; Posey, J.E.; Coban Akdemir, Z.; Pehlivan, D.; Harel, T.; Jhangiani, S.N.; Bayram, Y.; Song, X.; Bahrambeigi, V.; Yuregir, O.O.; et al. Phenotypic Expansion Illuminates Multilocus Pathogenic Variation. Genet. Med. 2018, 20, 1528–1537. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef]
- Lal, D.; Neubauer, B.A.; Toliat, M.R.; Altmüller, J.; Thiele, H.; Nürnberg, P.; Kamrath, C.; Schänzer, A.; Sander, T.; Hahn, A.; et al. Increased Probability of Co-Occurrence of Two Rare Diseases in Consanguineous Families and Resolution of a Complex Phenotype by Next Generation Sequencing. PLoS ONE 2016, 11, e0146040. [Google Scholar] [CrossRef]
- Matis, T.; Michaud, V.; Van-Gils, J.; Raclet, V.; Plaisant, C.; Fergelot, P.; Lasseaux, E.; Arveiler, B.; Trimouille, A. Triple Diagnosis of Wiedemann-Steiner, Waardenburg and DLG3-Related Intellectual Disability Association Found by WES: A Case Report. J. Gene Med. 2020, 22, e3197. [Google Scholar] [CrossRef]
- Li, Y.; Salfelder, A.; Schwab, K.O.; Grünert, S.C.; Velten, T.; Lütjohann, D.; Villavicencio-Lorini, P.; Matysiak-Scholze, U.; Zabel, B.; Köttgen, A.; et al. Against All Odds: Blended Phenotypes of Three Single-Gene Defects. Eur. J. Hum. Genet. 2016, 24, 1274–1279. [Google Scholar] [CrossRef][Green Version]
- Jehee, F.S.; de Oliveira, V.T.; Gurgel-Giannetti, J.; Pietra, R.X.; Rubatino, F.V.M.; Carobin, N.V.; Vianna, G.S.; de Freitas, M.L.; Fernandes, K.S.; Ribeiro, B.S.V.; et al. Dual Molecular Diagnosis Contributes to Atypical Prader-Willi Phenotype in Monozygotic Twins. Am. J. Med. Genet. A 2017, 173, 2451–2455. [Google Scholar] [CrossRef]
- Jourdon, A.; Fasching, L.; Scuderi, S.; Abyzov, A.; Vaccarino, F.M. The Role of Somatic Mosaicism in Brain Disease. Curr. Opin. Genet Dev. 2020, 65, 84–90. [Google Scholar] [CrossRef]
- Freed, D.; Pevsner, J. The Contribution of Mosaic Variants to Autism Spectrum Disorder. PLoS Genet 2016, 12, e1006245. [Google Scholar] [CrossRef]
- Dou, Y.; Yang, X.; Li, Z.; Wang, S.; Zhang, Z.; Ye, A.Y.; Yan, L.; Yang, C.; Wu, Q.; Li, J.; et al. Postzygotic Single-Nucleotide Mosaicisms Contribute to the Etiology of Autism Spectrum Disorder and Autistic Traits and the Origin of Mutations. Hum. Mutat. 2017, 38, 1002–1013. [Google Scholar] [CrossRef]
- Ansari, M.; Poke, G.; Ferry, Q.; Williamson, K.; Aldridge, R.; Meynert, A.M.; Bengani, H.; Chan, C.Y.; Kayserili, H.; Avci, S.; et al. Genetic Heterogeneity in Cornelia de Lange Syndrome (CdLS) and CdLS-like Phenotypes with Observed and Predicted Levels of Mosaicism. J. Med. Genet. 2014, 51, 659–668. [Google Scholar] [CrossRef]
- Krawczynska, N.; Wierzba, J.; Wasag, B. Genetic Mosaicism in a Group of Patients with Cornelia de Lange Syndrome. Front Pediatr 2019, 7, 203. [Google Scholar] [CrossRef]
- Stosser, M.B.; Lindy, A.S.; Butler, E.; Retterer, K.; Piccirillo-Stosser, C.M.; Richard, G.; McKnight, D.A. High Frequency of Mosaic Pathogenic Variants in Genes Causing Epilepsy-Related Neurodevelopmental Disorders. Genet. Med. 2018, 20, 403–410. [Google Scholar] [CrossRef]
- D’Gama, A.M.; Walsh, C.A. Somatic Mosaicism and Neurodevelopmental Disease. Nat. Neurosci. 2018, 21, 1504–1514. [Google Scholar] [CrossRef]
- Potter, H.; Chial, H.J.; Caneus, J.; Elos, M.; Elder, N.; Borysov, S.; Granic, A. Chromosome Instability and Mosaic Aneuploidy in Neurodegenerative and Neurodevelopmental Disorders. Front. Genet. 2019, 10, 1092. [Google Scholar] [CrossRef]
- Rohrback, S.; Siddoway, B.; Liu, C.S.; Chun, J. Genomic Mosaicism in the Developing and Adult Brain. Dev. Neurobiol. 2018, 78, 1026–1048. [Google Scholar] [CrossRef]
- D’Gama, A.M.; Woodworth, M.B.; Hossain, A.A.; Bizzotto, S.; Hatem, N.E.; LaCoursiere, C.M.; Najm, I.; Ying, Z.; Yang, E.; Barkovich, A.J.; et al. Somatic Mutations Activating the MTOR Pathway in Dorsal Telencephalic Progenitors Cause a Continuum of Cortical Dysplasias. Cell Rep. 2017, 21, 3754–3766. [Google Scholar] [CrossRef]
- McNulty, S.N.; Evenson, M.J.; Corliss, M.M.; Love-Gregory, L.D.; Schroeder, M.C.; Cao, Y.; Lee, Y.-S.; Drolet, B.A.; Neidich, J.A.; Cottrell, C.E.; et al. Diagnostic Utility of Next-Generation Sequencing for Disorders of Somatic Mosaicism: A Five-Year Cumulative Cohort. Am. J. Hum. Genet. 2019, 105, 734–746. [Google Scholar] [CrossRef]
- Wright, C.F.; Prigmore, E.; Rajan, D.; Handsaker, J.; McRae, J.; Kaplanis, J.; Fitzgerald, T.W.; FitzPatrick, D.R.; Firth, H.V.; Hurles, M.E. Clinically-Relevant Postzygotic Mosaicism in Parents and Children with Developmental Disorders in Trio Exome Sequencing Data. Nat. Commun. 2019, 10, 2985. [Google Scholar] [CrossRef]
- Chang, F.; Liu, L.; Fang, E.; Zhang, G.; Chen, T.; Cao, K.; Li, Y.; Li, M.M. Molecular Diagnosis of Mosaic Overgrowth Syndromes Using a Custom-Designed Next-Generation Sequencing Panel. J. Mol. Diagn. 2017, 19, 613–624. [Google Scholar] [CrossRef]
- Krawczynska, N.; Kuzniacka, A.; Wierzba, J.; Parenti, I.; Kaiser, F.J.; Wasag, B. Mosaic Intronic NIPBL Variant in a Family With Cornelia de Lange Syndrome. Front. Genet. 2018, 9, 255. [Google Scholar] [CrossRef]
- Theda, C.; Hwang, S.H.; Czajko, A.; Loke, Y.J.; Leong, P.; Craig, J.M. Quantitation of the Cellular Content of Saliva and Buccal Swab Samples. Sci. Rep. 2018, 8, 6944. [Google Scholar] [CrossRef]
- Ruggieri, M.; Praticò, A.D. Mosaic Neurocutaneous Disorders and Their Causes. Semin. Pediatr. Neurol. 2015, 22, 207–233. [Google Scholar] [CrossRef]
- Cao, Y.; Tokita, M.J.; Chen, E.S.; Ghosh, R.; Chen, T.; Feng, Y.; Gorman, E.; Gibellini, F.; Ward, P.A.; Braxton, A.; et al. A Clinical Survey of Mosaic Single Nucleotide Variants in Disease-Causing Genes Detected by Exome Sequencing. Genome Med. 2019, 11, 48. [Google Scholar] [CrossRef]
- Retterer, K.; Juusola, J.; Cho, M.T.; Vitazka, P.; Millan, F.; Gibellini, F.; Vertino-Bell, A.; Smaoui, N.; Neidich, J.; Monaghan, K.G.; et al. Clinical Application of Whole-Exome Sequencing across Clinical Indications. Genet. Med. 2016, 18, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Gold, H.D.; Luquette, L.J.; Park, P.J. Detecting Somatic Mutations in Normal Cells. Trends Genet. 2018, 34, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bae, T.; Thorpe, J.; Sherman, M.A.; Jones, A.G.; Cho, S.; Daily, K.; Dou, Y.; Ganz, J.; Galor, A.; et al. Comprehensive Identification of Somatic Nucleotide Variants in Human Brain Tissue. Genome Biol. 2021, 22, 92. [Google Scholar] [CrossRef] [PubMed]
- King, D.A.; Sifrim, A.; Fitzgerald, T.W.; Rahbari, R.; Hobson, E.; Homfray, T.; Mansour, S.; Mehta, S.G.; Shehla, M.; Tomkins, S.E.; et al. Detection of Structural Mosaicism from Targeted and Whole-Genome Sequencing Data. Genome Res. 2017, 27, 1704–1714. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.Y.; Zhang, Z.; Ye, A.Y.; Dou, Y.; Yan, L.; Yang, X.; Zhang, Y.; Wei, L. MosaicHunter: Accurate Detection of Postzygotic Single-Nucleotide Mosaicism through next-Generation Sequencing of Unpaired, Trio, and Paired Samples. Nucleic Acids Res. 2017, 45, e76. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Kwon, M.; Rodin, R.E.; Cortés-Ciriano, I.; Doan, R.; Luquette, L.J.; Galor, A.; Bohrson, C.; Walsh, C.A.; Park, P.J. Accurate Detection of Mosaic Variants in Sequencing Data without Matched Controls. Nat. Biotechnol. 2020, 38, 314–319. [Google Scholar] [CrossRef]
- Breuss, M.W.; Antaki, D.; George, R.D.; Kleiber, M.; James, K.N.; Ball, L.L.; Hong, O.; Mitra, I.; Yang, X.; Wirth, S.A.; et al. Autism Risk in Offspring Can Be Assessed through Quantification of Male Sperm Mosaicism. Nat. Med. 2020, 26, 143–150. [Google Scholar] [CrossRef]
- Salinas, V.; Vega, P.; Marsili, L.; Pérez-Maturo, J.; Martínez, N.; Zavala, L.; González-Morón, D.; Medina, N.; Rodriguez-Quiroga, S.A.; Amartino, H.; et al. The Odyssey of Complex Neurogenetic Disorders: From Undetermined to Positive. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 876–884. [Google Scholar] [CrossRef]
- Tan, N.B.; Stapleton, R.; Stark, Z.; Delatycki, M.B.; Yeung, A.; Hunter, M.F.; Amor, D.J.; Brown, N.J.; Stutterd, C.A.; McGillivray, G.; et al. Evaluating Systematic Reanalysis of Clinical Genomic Data in Rare Disease from Single Center Experience and Literature Review. Mol. Genet. Genomic Med. 2020, 8, e1508. [Google Scholar] [CrossRef]
- Burdick, K.J.; Cogan, J.D.; Rives, L.C.; Robertson, A.K.; Koziura, M.E.; Brokamp, E.; Duncan, L.; Hannig, V.; Pfotenhauer, J.; Vanzo, R.; et al. Limitations of Exome Sequencing in Detecting Rare and Undiagnosed Diseases. Am. J. Med. Genet. A 2020, 182, 1400–1406. [Google Scholar] [CrossRef]
- Deignan, J.L.; Chung, W.K.; Kearney, H.M.; Monaghan, K.G.; Rehder, C.W.; Chao, E.C. ACMG Laboratory Quality Assurance Committee Points to Consider in the Reevaluation and Reanalysis of Genomic Test Results: A Statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2019, 21, 1267–1270. [Google Scholar] [CrossRef]
- Shashi, V.; Schoch, K.; Spillmann, R.; Cope, H.; Tan, Q.K.-G.; Walley, N.; Pena, L.; McConkie-Rosell, A.; Jiang, Y.-H.; Stong, N.; et al. A Comprehensive Iterative Approach Is Highly Effective in Diagnosing Individuals Who Are Exome Negative. Genet. Med. 2019, 21, 161–172. [Google Scholar] [CrossRef]
- Need, A.C.; Shashi, V.; Schoch, K.; Petrovski, S.; Goldstein, D.B. The Importance of Dynamic Re-Analysis in Diagnostic Whole Exome Sequencing. J. Med. Genet. 2017, 54, 155–156. [Google Scholar] [CrossRef]
- Li, H.; Dawood, M.; Khayat, M.M.; Farek, J.R.; Jhangiani, S.N.; Khan, Z.M.; Mitani, T.; Coban-Akdemir, Z.; Lupski, J.R.; Venner, E.; et al. Exome Variant Discrepancies Due to Reference-Genome Differences. Am. J. Hum. Genet. 2021, 108, 1239–1250. [Google Scholar] [CrossRef]
- Baker, S.W.; Murrell, J.R.; Nesbitt, A.I.; Pechter, K.B.; Balciuniene, J.; Zhao, X.; Yu, Z.; Denenberg, E.H.; DeChene, E.T.; Wilkens, A.B.; et al. Automated Clinical Exome Reanalysis Reveals Novel Diagnoses. J. Mol. Diagn. 2019, 21, 38–48. [Google Scholar] [CrossRef]
- Salfati, E.L.; Spencer, E.G.; Topol, S.E.; Muse, E.D.; Rueda, M.; Lucas, J.R.; Wagner, G.N.; Campman, S.; Topol, E.J.; Torkamani, A. Re-Analysis of Whole-Exome Sequencing Data Uncovers Novel Diagnostic Variants and Improves Molecular Diagnostic Yields for Sudden Death and Idiopathic Diseases. Genome Med. 2019, 11, 83. [Google Scholar] [CrossRef]
- Sobreira, N.; Schiettecatte, F.; Valle, D.; Hamosh, A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015, 36, 928–930. [Google Scholar] [CrossRef]
- Schaefer, J.; Lehne, M.; Schepers, J.; Prasser, F.; Thun, S. The Use of Machine Learning in Rare Diseases: A Scoping Review. Orphanet J. Rare Dis. 2020, 15, 145. [Google Scholar] [CrossRef]
- Sundaram, L.; Gao, H.; Padigepati, S.R.; McRae, J.F.; Li, Y.; Kosmicki, J.A.; Fritzilas, N.; Hakenberg, J.; Dutta, A.; Shon, J.; et al. Predicting the Clinical Impact of Human Mutation with Deep Neural Networks. Nat. Genet. 2018, 50, 1161–1170. [Google Scholar] [CrossRef]
- Dragusin, R.; Petcu, P.; Lioma, C.; Larsen, B.; Jørgensen, H.L.; Cox, I.J.; Hansen, L.K.; Ingwersen, P.; Winther, O. FindZebra: A Search Engine for Rare Diseases. Int. J. Med. Inform. 2013, 82, 528–538. [Google Scholar] [CrossRef]
- Dragusin, R.; Petcu, P.; Lioma, C.; Larsen, B.; Jørgensen, H.L.; Cox, I.J.; Hansen, L.K.; Ingwersen, P.; Winther, O. Specialized Tools Are Needed When Searching the Web for Rare Disease Diagnoses. Rare Dis. 2013, 1, e25001. [Google Scholar] [CrossRef][Green Version]
- Fujiwara, T.; Yamamoto, Y.; Kim, J.-D.; Buske, O.; Takagi, T. PubCaseFinder: A Case-Report-Based, Phenotype-Driven Differential-Diagnosis System for Rare Diseases. Am. J. Hum. Genet. 2018, 103, 389–399. [Google Scholar] [CrossRef]
- Chen, T.; Chen, Y.; Yuan, M.; Gerstein, M.; Li, T.; Liang, H.; Froehlich, T.; Lu, L. The Development of a Practical Artificial Intelligence Tool for Diagnosing and Evaluating Autism Spectrum Disorder: Multicenter Study. JMIR Med. Inform. 2020, 8, e15767. [Google Scholar] [CrossRef]
- Robinson, P.N.; Köhler, S.; Oellrich, A.; Sanger Mouse Genetics Project; Wang, K.; Mungall, C.J.; Lewis, S.E.; Washington, N.; Bauer, S.; Seelow, D.; et al. Improved Exome Prioritization of Disease Genes through Cross-Species Phenotype Comparison. Genome Res. 2014, 24, 340–348. [Google Scholar] [CrossRef]
- Cipriani, V.; Pontikos, N.; Arno, G.; Sergouniotis, P.I.; Lenassi, E.; Thawong, P.; Danis, D.; Michaelides, M.; Webster, A.R.; Moore, A.T.; et al. An Improved Phenotype-Driven Tool for Rare Mendelian Variant Prioritization: Benchmarking Exomiser on Real Patient Whole-Exome Data. Genes 2020, 11, E460. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, K.; Bustamante, C.D.; Ma, X.; Wong, W.H. Xrare: A Machine Learning Method Jointly Modeling Phenotypes and Genetic Evidence for Rare Disease Diagnosis. Genet. Med. 2019, 21, 2126–2134. [Google Scholar] [CrossRef]
- Toro, C.; Hori, R.T.; Malicdan, M.C.V.; Tifft, C.J.; Goldstein, A.; Gahl, W.A.; Adams, D.R.; Fauni, H.B.; Wolfe, L.A.; Xiao, J.; et al. A Recurrent de Novo Missense Mutation in UBTF Causes Developmental Neuroregression. Hum. Mol. Genet. 2018, 27, 691–705. [Google Scholar] [CrossRef]
- Ji, J.; Shen, L.; Bootwalla, M.; Quindipan, C.; Tatarinova, T.; Maglinte, D.T.; Buckley, J.; Raca, G.; Saitta, S.C.; Biegel, J.A.; et al. A Semiautomated Whole-Exome Sequencing Workflow Leads to Increased Diagnostic Yield and Identification of Novel Candidate Variants. Cold Spring Harb. Mol. Case Stud. 2019, 5, a003756. [Google Scholar] [CrossRef]
- Basel-Salmon, L.; Orenstein, N.; Markus-Bustani, K.; Ruhrman-Shahar, N.; Kilim, Y.; Magal, N.; Hubshman, M.W.; Bazak, L. Improved Diagnostics by Exome Sequencing Following Raw Data Reevaluation by Clinical Geneticists Involved in the Medical Care of the Individuals Tested. Genet. Med. 2019, 21, 1443–1451. [Google Scholar] [CrossRef]
- Quaio, C.R.D.C.; Moreira, C.M.; Novo-Filho, G.M.; Sacramento-Bobotis, P.R.; Groenner Penna, M.; Perazzio, S.F.; Dutra, A.P.; da Silva, R.A.; Santos, M.N.P.; de Arruda, V.Y.N.; et al. Diagnostic Power and Clinical Impact of Exome Sequencing in a Cohort of 500 Patients with Rare Diseases. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 955–964. [Google Scholar] [CrossRef]
- Reinstein, E.; Tzur, S.; Cohen, R.; Bormans, C.; Behar, D.M. Intellectual Disability and Non-Compaction Cardiomyopathy with a de Novo NONO Mutation Identified by Exome Sequencing. Eur. J. Hum. Genet. 2016, 24, 1635–1638. [Google Scholar] [CrossRef]
- Mighton, C.; Smith, A.C.; Mayers, J.; Tomaszewski, R.; Taylor, S.; Hume, S.; Agatep, R.; Spriggs, E.; Feilotter, H.E.; Semenuk, L.; et al. Data Sharing to Improve Concordance in Variant Interpretation across Laboratories: Results from the Canadian Open Genetics Repository. J. Med. Genet. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gafner, M.; Michelson, M.; Yosovich, K.; Blumkin, L.; Lerman-Sagie, T.; Lev, D. Infantile Onset Progressive Cerebellar Atrophy and Anterior Horn Cell Degeneration-A Novel Phenotype Associated with Mutations in the PLA2G6 Gene. Eur. J. Med. Genet. 2020, 63, 103801. [Google Scholar] [CrossRef] [PubMed]
- Weiss, K.; Kurolap, A.; Paperna, T.; Mory, A.; Steinberg, M.; Hershkovitz, T.; Ekhilevitch, N.; Baris, H.N. Rare Disease Diagnostics: A Single-Center Experience and Lessons Learnt. Rambam. Maimonides Med. J. 2018, 9, e10018. [Google Scholar] [CrossRef] [PubMed]
- Kremer, L.S.; Bader, D.M.; Mertes, C.; Kopajtich, R.; Pichler, G.; Iuso, A.; Haack, T.B.; Graf, E.; Schwarzmayr, T.; Terrile, C.; et al. Genetic Diagnosis of Mendelian Disorders via RNA Sequencing. Nat. Commun. 2017, 8, 15824. [Google Scholar] [CrossRef] [PubMed]
- Dunn, P.; Prigatano, G.P.; Szelinger, S.; Roth, J.; Siniard, A.L.; Claasen, A.M.; Richholt, R.F.; De Both, M.; Corneveaux, J.J.; Moskowitz, A.M.; et al. A de Novo Splice Site Mutation in CASK Causes FG Syndrome-4 and Congenital Nystagmus. Am. J. Med. Genet. A 2017, 173, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Szelinger, S.; Malenica, I.; Corneveaux, J.J.; Siniard, A.L.; Kurdoglu, A.A.; Ramsey, K.M.; Schrauwen, I.; Trent, J.M.; Narayanan, V.; Huentelman, M.J.; et al. Characterization of X Chromosome Inactivation Using Integrated Analysis of Whole-Exome and MRNA Sequencing. PLoS ONE 2014, 9, e113036. [Google Scholar] [CrossRef]
- Schrauwen, I.; Szelinger, S.; Siniard, A.L.; Corneveaux, J.J.; Kurdoglu, A.; Richholt, R.; De Both, M.; Malenica, I.; Swaminathan, S.; Rangasamy, S.; et al. A De Novo Mutation in TEAD1 Causes Non-X-Linked Aicardi Syndrome. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3896–3904. [Google Scholar] [CrossRef][Green Version]
- Plenge, R.M.; Stevenson, R.A.; Lubs, H.A.; Schwartz, C.E.; Willard, H.F. Skewed X-Chromosome Inactivation Is a Common Feature of X-Linked Mental Retardation Disorders. Am. J. Hum. Genet. 2002, 71, 168–173. [Google Scholar] [CrossRef]
- Frésard, L.; Smail, C.; Ferraro, N.M.; Teran, N.A.; Li, X.; Smith, K.S.; Bonner, D.; Kernohan, K.D.; Marwaha, S.; Zappala, Z.; et al. Identification of Rare-Disease Genes Using Blood Transcriptome Sequencing and Large Control Cohorts. Nat. Med. 2019, 25, 911–919. [Google Scholar] [CrossRef]
- Aygun, D.; Bjornsson, H.T. Clinical Epigenetics: A Primer for the Practitioner. Dev. Med. Child Neurol. 2020, 62, 192–200. [Google Scholar] [CrossRef]
- Mossink, B.; Negwer, M.; Schubert, D.; Nadif Kasri, N. The Emerging Role of Chromatin Remodelers in Neurodevelopmental Disorders: A Developmental Perspective. Cell Mol. Life Sci. 2021, 78, 2517–2563. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; Groupe DI France; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.-C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020, 106, 356–370. [Google Scholar] [CrossRef]
- Godler, D.E.; Amor, D.J. DNA Methylation Analysis for Screening and Diagnostic Testing in Neurodevelopmental Disorders. Essays Biochem. 2019, 63, 785–795. [Google Scholar] [CrossRef]
- Haghshenas, S.; Bhai, P.; Aref-Eshghi, E.; Sadikovic, B. Diagnostic Utility of Genome-Wide DNA Methylation Analysis in Mendelian Neurodevelopmental Disorders. Int. J. Mol. Sci. 2020, 21, E9303. [Google Scholar] [CrossRef]
- Sadikovic, B.; Levy, M.A.; Kerkhof, J.; Aref-Eshghi, E.; Schenkel, L.; Stuart, A.; McConkey, H.; Henneman, P.; Venema, A.; Schwartz, C.E.; et al. Clinical Epigenomics: Genome-Wide DNA Methylation Analysis for the Diagnosis of Mendelian Disorders. Genet. Med. 2021, 23, 1065–1074. [Google Scholar] [CrossRef]
- Gackowski, D.; Zarakowska, E.; Starczak, M.; Modrzejewska, M.; Olinski, R. Tissue-Specific Differences in DNA Modifications (5-Hydroxymethylcytosine, 5-Formylcytosine, 5-Carboxylcytosine and 5-Hydroxymethyluracil) and Their Interrelationships. PLoS ONE 2015, 10, e0144859. [Google Scholar] [CrossRef]
- Wen, L.; Tang, F. Genomic Distribution and Possible Functions of DNA Hydroxymethylation in the Brain. Genomics 2014, 104, 341–346. [Google Scholar] [CrossRef]
- Melo, U.S.; Schöpflin, R.; Acuna-Hidalgo, R.; Mensah, M.A.; Fischer-Zirnsak, B.; Holtgrewe, M.; Klever, M.-K.; Türkmen, S.; Heinrich, V.; Pluym, I.D.; et al. Hi-C Identifies Complex Genomic Rearrangements and TAD-Shuffling in Developmental Diseases. Am. J. Hum. Genet. 2020, 106, 872–884. [Google Scholar] [CrossRef]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.P.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Manes, N.P.; Nita-Lazar, A. Application of Targeted Mass Spectrometry in Bottom-up Proteomics for Systems Biology Research. J. Proteomics 2018, 189, 75–90. [Google Scholar] [CrossRef]
- Boersema, P.J.; Kahraman, A.; Picotti, P. Proteomics beyond Large-Scale Protein Expression Analysis. Curr. Opin. Biotechnol. 2015, 34, 162–170. [Google Scholar] [CrossRef]
- Meng, W.; Huan, Y.; Gao, Y. Urinary Proteome Profiling for Children with Autism Using Data-Independent Acquisition Proteomics. Transl. Pediatr. 2021, 10, 1765–1778. [Google Scholar] [CrossRef]
- Mesleh, A.G.; Abdulla, S.A.; El-Agnaf, O. Paving the Way toward Personalized Medicine: Current Advances and Challenges in Multi-OMICS Approach in Autism Spectrum Disorder for Biomarkers Discovery and Patient Stratification. J. Pers. Med. 2021, 11, 41. [Google Scholar] [CrossRef]
- Murtaza, N.; Uy, J.; Singh, K.K. Emerging Proteomic Approaches to Identify the Underlying Pathophysiology of Neurodevelopmental and Neurodegenerative Disorders. Mol. Autism. 2020, 11, 27. [Google Scholar] [CrossRef]
- Mingirulli, N.; Pyle, A.; Hathazi, D.; Alston, C.L.; Kohlschmidt, N.; O’Grady, G.; Waddell, L.; Evesson, F.; Cooper, S.B.T.; Turner, C.; et al. Clinical Presentation and Proteomic Signature of Patients with TANGO2 Mutations. J. Inherit. Metab. Dis. 2020, 43, 297–308. [Google Scholar] [CrossRef]
- Gungor, S.; Oktay, Y.; Hiz, S.; Aranguren-Ibáñez, Á.; Kalafatcilar, I.; Yaramis, A.; Karaca, E.; Yis, U.; Sonmezler, E.; Ekinci, B.; et al. Autosomal Recessive Variants in TUBGCP2 Alter the γ-Tubulin Ring Complex Leading to Neurodevelopmental Disease. iScience 2021, 24, 101948. [Google Scholar] [CrossRef]
- Roos, A.; Thompson, R.; Horvath, R.; Lochmüller, H.; Sickmann, A. Intersection of Proteomics and Genomics to “Solve the Unsolved” in Rare Disorders Such as Neurodegenerative and Neuromuscular Diseases. Proteomics Clin. Appl. 2018, 12. [Google Scholar] [CrossRef]
- Buccitelli, C.; Selbach, M. MRNAs, Proteins and the Emerging Principles of Gene Expression Control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef]
- Kustatscher, G.; Grabowski, P.; Rappsilber, J. Pervasive Coexpression of Spatially Proximal Genes Is Buffered at the Protein Level. Mol. Syst. Biol. 2017, 13, 937. [Google Scholar] [CrossRef]
- Liu, Y.; Borel, C.; Li, L.; Müller, T.; Williams, E.G.; Germain, P.-L.; Buljan, M.; Sajic, T.; Boersema, P.J.; Shao, W.; et al. Systematic Proteome and Proteostasis Profiling in Human Trisomy 21 Fibroblast Cells. Nat. Commun. 2017, 8, 1212. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wu, L.; Liao, H.; Lu, X.; Zhang, X.; Kuang, X.; Yang, L. Clinical and Genetic Analysis of Six Chinese Children with Poirier-Bienvenu Neurodevelopmental Syndrome Caused by CSNK2B Mutation. Neurogenetics 2021, 22, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.D.; Pappan, K.L.; Donti, T.; Delgado, M.R.; Shinawi, M.; Pearson, T.S.; Lalani, S.R.; Craigen, W.E.; Sutton, V.R.; Evans, A.M.; et al. 2-Pyrrolidinone and Succinimide as Clinical Screening Biomarkers for GABA-Transaminase Deficiency: Anti-Seizure Medications Impact Accurate Diagnosis. Front. Neurosci. 2019, 13, 394. [Google Scholar] [CrossRef] [PubMed]
- Kerkhofs, M.H.P.M.; Haijes, H.A.; Willemsen, A.M.; van Gassen, K.L.I.; van der Ham, M.; Gerrits, J.; de Sain-van der Velden, M.G.M.; Prinsen, H.C.M.T.; van Deutekom, H.W.M.; van Hasselt, P.M.; et al. Cross-Omics: Integrating Genomics with Metabolomics in Clinical Diagnostics. Metabolites 2020, 10, E206. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for Laboratory Diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Lussu, M.; Noto, A.; Masili, A.; Rinaldi, A.C.; Dessì, A.; De Angelis, M.; De Giacomo, A.; Fanos, V.; Atzori, L.; Francavilla, R. The Urinary 1 H-NMR Metabolomics Profile of an Italian Autistic Children Population and Their Unaffected Siblings. Autism Res. 2017, 10, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Shyr, C.; Ross, C.J.; Horvath, G.A.; Salvarinova, R.; Ye, X.C.; Zhang, L.-H.; Bhavsar, A.P.; Lee, J.J.Y.; Drögemöller, B.I.; et al. Exome Sequencing and the Management of Neurometabolic Disorders. N. Engl. J. Med. 2016, 374, 2246–2255. [Google Scholar] [CrossRef]
- Sarigiannis, D.A.; Papaioannou, N.; Handakas, E.; Anesti, O.; Polanska, K.; Hanke, W.; Salifoglou, A.; Gabriel, C.; Karakitsios, S. Neurodevelopmental Exposome: The Effect of in Utero Co-Exposure to Heavy Metals and Phthalates on Child Neurodevelopment. Environ. Res. 2021, 197, 110949. [Google Scholar] [CrossRef]
- Bardanzellu, F.; Fanos, V. How Could Metabolomics Change Pediatric Health? Ital. J. Pediatr. 2020, 46, 37. [Google Scholar] [CrossRef]
- Orozco, J.S.; Hertz-Picciotto, I.; Abbeduto, L.; Slupsky, C.M. Metabolomics Analysis of Children with Autism, Idiopathic-Developmental Delays, and Down Syndrome. Transl. Psychiatry 2019, 9, 243. [Google Scholar] [CrossRef]
- Sotelo-Orozco, J.; Abbeduto, L.; Hertz-Picciotto, I.; Slupsky, C.M. Association between Plasma Metabolites and Psychometric Scores Among Children With Developmental Disabilities: Investigating Sex-Differences. Front. Psychiatry 2020, 11, 579538. [Google Scholar] [CrossRef]
- Neul, J.L.; Skinner, S.A.; Annese, F.; Lane, J.; Heydemann, P.; Jones, M.; Kaufmann, W.E.; Glaze, D.G.; Percy, A.K. Metabolic Signatures Differentiate Rett Syndrome From Unaffected Siblings. Front. Integr. Neurosci. 2020, 14, 7. [Google Scholar] [CrossRef]
- Coene, K.L.M.; Kluijtmans, L.A.J.; van der Heeft, E.; Engelke, U.F.H.; de Boer, S.; Hoegen, B.; Kwast, H.J.T.; van de Vorst, M.; Huigen, M.C.D.G.; Keularts, I.M.L.W.; et al. Next-Generation Metabolic Screening: Targeted and Untargeted Metabolomics for the Diagnosis of Inborn Errors of Metabolism in Individual Patients. J. Inherit. Metab. Dis. 2018, 41, 337–353. [Google Scholar] [CrossRef]
- Lee, J.J.Y.; Wasserman, W.W.; Hoffmann, G.F.; van Karnebeek, C.D.M.; Blau, N. Knowledge Base and Mini-Expert Platform for the Diagnosis of Inborn Errors of Metabolism. Genet. Med. 2018, 20, 151–158. [Google Scholar] [CrossRef]
- Márquez-Caraveo, M.E.; Ibarra-González, I.; Rodríguez-Valentín, R.; Ramírez-García, M.Á.; Pérez-Barrón, V.; Lazcano-Ponce, E.; Vela-Amieva, M. Brief Report: Delayed Diagnosis of Treatable Inborn Errors of Metabolism in Children with Autism and Other Neurodevelopmental Disorders. J. Autism Dev. Disord. 2021, 51, 2124–2131. [Google Scholar] [CrossRef]
- Saudubray, J.-M.; Garcia-Cazorla, A. An Overview of Inborn Errors of Metabolism Affecting the Brain: From Neurodevelopment to Neurodegenerative Disorders. Dialogues Clin. Neurosci. 2018, 20, 301–325. [Google Scholar] [CrossRef]
- Hoytema van Konijnenburg, E.M.M.; Wortmann, S.B.; Koelewijn, M.J.; Tseng, L.A.; Houben, R.; Stöckler-Ipsiroglu, S.; Ferreira, C.R.; van Karnebeek, C.D.M. Treatable Inherited Metabolic Disorders Causing Intellectual Disability: 2021 Review and Digital App. Orphanet. J. Rare Dis. 2021, 16, 170. [Google Scholar] [CrossRef]
- Ortigoza-Escobar, J.D. A Proposed Diagnostic Algorithm for Inborn Errors of Metabolism Presenting With Movements Disorders. Front Neurol 2020, 11, 582160. [Google Scholar] [CrossRef]
- GTEx Consortium the Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [CrossRef]
- Eze, U.C.; Bhaduri, A.; Haeussler, M.; Nowakowski, T.J.; Kriegstein, A.R. Single-Cell Atlas of Early Human Brain Development Highlights Heterogeneity of Human Neuroepithelial Cells and Early Radial Glia. Nat. Neurosci. 2021, 24, 584–594. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Bhaduri, A.; Pollen, A.A.; Alvarado, B.; Mostajo-Radji, M.A.; Di Lullo, E.; Haeussler, M.; Sandoval-Espinosa, C.; Liu, S.J.; Velmeshev, D.; et al. Spatiotemporal Gene Expression Trajectories Reveal Developmental Hierarchies of the Human Cortex. Science 2017, 358, 1318–1323. [Google Scholar] [CrossRef]
- Zeisel, A.; Hochgerner, H.; Lönnerberg, P.; Johnsson, A.; Memic, F.; van der Zwan, J.; Häring, M.; Braun, E.; Borm, L.E.; La Manno, G.; et al. Molecular Architecture of the Mouse Nervous System. Cell 2018, 174, 999–1014.e22. [Google Scholar] [CrossRef]
- Sunkin, S.M.; Ng, L.; Lau, C.; Dolbeare, T.; Gilbert, T.L.; Thompson, C.L.; Hawrylycz, M.; Dang, C. Allen Brain Atlas: An Integrated Spatio-Temporal Portal for Exploring the Central Nervous System. Nucleic Acids Res. 2013, 41, D996–D1008. [Google Scholar] [CrossRef]
- Speir, M.L.; Bhaduri, A.; Markov, N.S.; Moreno, P.; Nowakowski, T.J.; Papatheodorou, I.; Pollen, A.A.; Raney, B.J.; Seninge, L.; Kent, W.J.; et al. UCSC Cell Browser: Visualize Your Single-Cell Data. Bioinformatics 2021, btab503. [Google Scholar] [CrossRef]
- Stark, C.; Breitkreutz, B.-J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A General Repository for Interaction Datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A Database of Reactions, Pathways and Biological Processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef]
- Schrauwen, I.; Szelinger, S.; Siniard, A.L.; Kurdoglu, A.; Corneveaux, J.J.; Malenica, I.; Richholt, R.; Van Camp, G.; De Both, M.; Swaminathan, S.; et al. A Frame-Shift Mutation in CAV1 Is Associated with a Severe Neonatal Progeroid and Lipodystrophy Syndrome. PLoS ONE 2015, 10, e0131797. [Google Scholar] [CrossRef][Green Version]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Ritchie, G.R.S.; Dunham, I.; Zeggini, E.; Flicek, P. Functional Annotation of Non-Coding Sequence Variants. Nat. Methods 2014, 11, 294–296. [Google Scholar] [CrossRef]
- Ionita-Laza, I.; McCallum, K.; Xu, B.; Buxbaum, J.D. A Spectral Approach Integrating Functional Genomic Annotations for Coding and Noncoding Variants. Nat. Genet. 2016, 48, 214–220. [Google Scholar] [CrossRef]
- Zhou, J.; Troyanskaya, O.G. Predicting Effects of Noncoding Variants with Deep Learning–Based Sequence Model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.; Shah, N.H.; Hripcsak, G. Deep Phenotyping: Embracing Complexity and Temporality-Towards Scalability, Portability, and Interoperability. J. Biomed. Inform. 2020, 105, 103433. [Google Scholar] [CrossRef] [PubMed]
- Yehia, L.; Eng, C. Largescale Population Genomics versus Deep Phenotyping: Brute Force or Elegant Pragmatism towards Precision Medicine. NPJ Genom. Med. 2019, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Pena, L.D.M.; Jiang, Y.-H.; Schoch, K.; Spillmann, R.C.; Walley, N.; Stong, N.; Rapisardo Horn, S.; Sullivan, J.A.; McConkie-Rosell, A.; Kansagra, S.; et al. Looking beyond the Exome: A Phenotype-First Approach to Molecular Diagnostic Resolution in Rare and Undiagnosed Diseases. Genet. Med. 2018, 20, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Girdea, M.; Dumitriu, S.; Fiume, M.; Bowdin, S.; Boycott, K.M.; Chénier, S.; Chitayat, D.; Faghfoury, H.; Meyn, M.S.; Ray, P.N.; et al. PhenoTips: Patient Phenotyping Software for Clinical and Research Use. Hum. Mutat. 2013, 34, 1057–1065. [Google Scholar] [CrossRef]
- Hamilton, C.M.; Strader, L.C.; Pratt, J.G.; Maiese, D.; Hendershot, T.; Kwok, R.K.; Hammond, J.A.; Huggins, W.; Jackman, D.; Pan, H.; et al. The PhenX Toolkit: Get the Most from Your Measures. Am. J. Epidemiol. 2011, 174, 253–260. [Google Scholar] [CrossRef]
- Philippakis, A.A.; Azzariti, D.R.; Beltran, S.; Brookes, A.J.; Brownstein, C.A.; Brudno, M.; Brunner, H.G.; Buske, O.J.; Carey, K.; Doll, C.; et al. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum. Mutat. 2015, 36, 915–921. [Google Scholar] [CrossRef]
- Mantere, T.; Kersten, S.; Hoischen, A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019, 10, 426. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Lee, H.; Darby, C.A.; Schatz, M.C. Piercing the Dark Matter: Bioinformatics of Long-Range Sequencing and Mapping. Nat. Rev. Genet. 2018, 19, 329–346. [Google Scholar] [CrossRef]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-Read Human Genome Sequencing and Its Applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef]
- Bowden, R.; Davies, R.W.; Heger, A.; Pagnamenta, A.T.; de Cesare, M.; Oikkonen, L.E.; Parkes, D.; Freeman, C.; Dhalla, F.; Patel, S.Y.; et al. Sequencing of Human Genomes with Nanopore Technology. Nat. Commun. 2019, 10, 1869. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and Challenges in Long-Read Sequencing Data Analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Loit, K.; Adamson, K.; Bahram, M.; Puusepp, R.; Anslan, S.; Kiiker, R.; Drenkhan, R.; Tedersoo, L. Relative Performance of MinION (Oxford Nanopore Technologies) versus Sequel (Pacific Biosciences) Third-Generation Sequencing Instruments in Identification of Agricultural and Forest Fungal Pathogens. Appl. Environ. Microbiol. 2019, 85, e01368–e01419. [Google Scholar] [CrossRef]
- Lei, M.; Liang, D.; Yang, Y.; Mitsuhashi, S.; Katoh, K.; Miyake, N.; Frith, M.C.; Wu, L.; Matsumoto, N. Long-Read DNA Sequencing Fully Characterized Chromothripsis in a Patient with Langer-Giedion Syndrome and Cornelia de Lange Syndrome-4. J. Hum. Genet. 2020, 65, 667–674. [Google Scholar] [CrossRef]
- Xie, Z.; Sun, C.; Zhang, S.; Liu, Y.; Yu, M.; Zheng, Y.; Meng, L.; Acharya, A.; Cornejo-Sanchez, D.M.; Wang, G.; et al. Long-Read Whole-Genome Sequencing for the Genetic Diagnosis of Dystrophinopathies. Ann. Clin. Transl. Neurol. 2020, 7, 2041–2046. [Google Scholar] [CrossRef]
- Ohori, S.; Tsuburaya, R.S.; Kinoshita, M.; Miyagi, E.; Mizuguchi, T.; Mitsuhashi, S.; Frith, M.C.; Matsumoto, N. Long-Read Whole-Genome Sequencing Identified a Partial MBD5 Deletion in an Exome-Negative Patient with Neurodevelopmental Disorder. J. Hum. Genet. 2021, 66, 697–705. [Google Scholar] [CrossRef]
- Hiatt, S.M.; Lawlor, J.M.J.; Handley, L.H.; Ramaker, R.C.; Rogers, B.B.; Partridge, E.C.; Boston, L.B.; Williams, M.; Plott, C.B.; Jenkins, J.; et al. Long-Read Genome Sequencing for the Molecular Diagnosis of Neurodevelopmental Disorders. HGG Adv. 2021, 2, 100023. [Google Scholar] [CrossRef]
- Elyanow, R.; Wu, H.-T.; Raphael, B.J. Identifying Structural Variants Using Linked-Read Sequencing Data. Bioinformatics 2018, 34, 353–360. [Google Scholar] [CrossRef]
- Garcia, S.; Williams, S.; Xu, A.W.; Herschleb, J.; Marks, P.; Stafford, D.; Church, D.M. Linked-Read Sequencing Resolves Complex Structural Variants. bioRxiv 2017, 231662. [Google Scholar] [CrossRef]
- Wang, O.; Chin, R.; Cheng, X.; Wu, M.K.Y.; Mao, Q.; Tang, J.; Sun, Y.; Anderson, E.; Lam, H.K.; Chen, D.; et al. Efficient and Unique Cobarcoding of Second-Generation Sequencing Reads from Long DNA Molecules Enabling Cost-Effective and Accurate Sequencing, Haplotyping, and de Novo Assembly. Genome Res. 2019, 29, 798–808. [Google Scholar] [CrossRef]
- Marks, P.; Garcia, S.; Barrio, A.M.; Belhocine, K.; Bernate, J.; Bharadwaj, R.; Bjornson, K.; Catalanotti, C.; Delaney, J.; Fehr, A.; et al. Resolving the Full Spectrum of Human Genome Variation Using Linked-Reads. Genome Res. 2019, 29, 635–645. [Google Scholar] [CrossRef]
- Onore, M.E.; Torella, A.; Musacchia, F.; D’Ambrosio, P.; Zanobio, M.; Del Vecchio Blanco, F.; Piluso, G.; Nigro, V. Linked-Read Whole Genome Sequencing Solves a Double DMD Gene Rearrangement. Genes 2021, 12, 133. [Google Scholar] [CrossRef]
- Barseghyan, H.; Tang, W.; Wang, R.T.; Almalvez, M.; Segura, E.; Bramble, M.S.; Lipson, A.; Douine, E.D.; Lee, H.; Délot, E.C.; et al. Next-Generation Mapping: A Novel Approach for Detection of Pathogenic Structural Variants with a Potential Utility in Clinical Diagnosis. Genome Med. 2017, 9, 90. [Google Scholar] [CrossRef]
- Mohr, D.W.; Naguib, A.; Weisenfeld, N.I.; Kumar, V.; Shah, P.; Church, D.M.; Jaffe, D.; Scott, A.F. Improved de Novo Genome Assembly: Linked-Read Sequencing Combined with Optical Mapping Produce a High Quality Mammalian Genome at Relatively Low Cost. bioRxiv 2017, 128348. [Google Scholar] [CrossRef]
- Cope, H.; Barseghyan, H.; Bhattacharya, S.; Fu, Y.; Hoppman, N.; Marcou, C.; Walley, N.; Rehder, C.; Deak, K.; Alkelai, A.; et al. Detection of a Mosaic CDKL5 Deletion and Inversion by Optical Genome Mapping Ends an Exhaustive Diagnostic Odyssey. Mol. Genet. Genomic Med. 2021, 9, e1665. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, M.; Qian, Y.; Yang, Y.; Sun, Y.; Liu, B.; Wang, L.; Dong, M. Identification of a Likely Pathogenic Structural Variation in the LAMA1 Gene by Bionano Optical Mapping. npj Genomic Medicine 2020, 5, 1–6. [Google Scholar] [CrossRef]
- Stence, A.A.; Thomason, J.G.; Pruessner, J.A.; Sompallae, R.R.; Snow, A.N.; Ma, D.; Moore, S.A.; Bossler, A.D. Validation of Optical Genome Mapping for the Molecular Diagnosis of Facioscapulohumeral Muscular Dystrophy. J. Mol. Diagn 2021, 23, 1506–1514. [Google Scholar] [CrossRef]
- Chintalaphani, S.R.; Pineda, S.S.; Deveson, I.W.; Kumar, K.R. An Update on the Neurological Short Tandem Repeat Expansion Disorders and the Emergence of Long-Read Sequencing Diagnostics. Acta Neuropathol. Commun. 2021, 9, 98. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadlubowska, M.K.; Schrauwen, I. Methods to Improve Molecular Diagnosis in Genomic Cold Cases in Pediatric Neurology. Genes 2022, 13, 333. https://doi.org/10.3390/genes13020333
Kadlubowska MK, Schrauwen I. Methods to Improve Molecular Diagnosis in Genomic Cold Cases in Pediatric Neurology. Genes. 2022; 13(2):333. https://doi.org/10.3390/genes13020333
Chicago/Turabian StyleKadlubowska, Magda K., and Isabelle Schrauwen. 2022. "Methods to Improve Molecular Diagnosis in Genomic Cold Cases in Pediatric Neurology" Genes 13, no. 2: 333. https://doi.org/10.3390/genes13020333
APA StyleKadlubowska, M. K., & Schrauwen, I. (2022). Methods to Improve Molecular Diagnosis in Genomic Cold Cases in Pediatric Neurology. Genes, 13(2), 333. https://doi.org/10.3390/genes13020333





